1 c3g5oC_
99.9
100
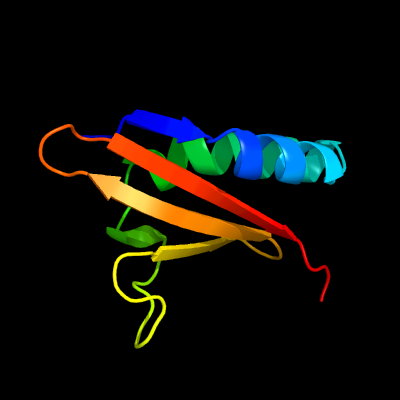
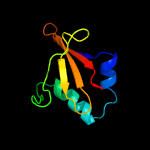
PDB header: toxin/antitoxinChain: C: PDB Molecule: uncharacterized protein rv2866;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
2 c2kheA_
99.9
35
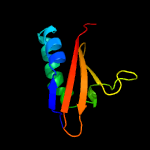
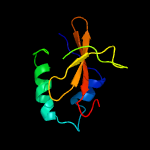
PDB header: hydrolaseChain: A: PDB Molecule: toxin-like protein;PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
3 d1wmia1
99.9
25
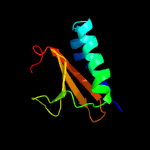
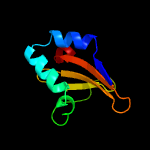
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like
4 c3kixy_
99.9
22
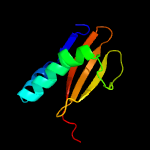
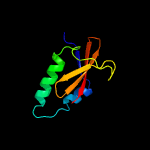
PDB header: ribosomeChain: Y: PDB Molecule: PDBTitle: structure of rele nuclease bound to the 70s ribosome (postcleavage2 state; part 3 of 4)
5 c3bpqD_
99.9
23
PDB header: toxinChain: D: PDB Molecule: toxin rele3;PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
6 c2otrA_
99.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hp0892;PDBTitle: solution structure of conserved hypothetical protein hp0892 from2 helicobacter pylori
7 c3oeiH_
99.3
29
PDB header: toxin, protein bindingChain: H: PDB Molecule: relk (toxin rv3358);PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
8 d2a6sa1
99.2
24
Fold: RelE-likeSuperfamily: RelE-likeFamily: YoeB/Txe-like
9 d1z8ma1
99.1
19
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like
10 c4q2uH_
98.9
16
PDB header: toxin/toxin repressorChain: H: PDB Molecule: mrna interferase yafq;PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
11 c5cegB_
98.7
20
PDB header: toxinChain: B: PDB Molecule: plasmid stabilization system;PDBTitle: x-ray structure of toxin/anti-toxin complex from mesorhizobium2 opportunistum
12 c5cw7H_
97.9
23
PDB header: toxinChain: H: PDB Molecule: plasmid stabilization protein pare;PDBTitle: crystal structure of the paaa2-pare2 antitoxin-toxin complex
13 c3kxeB_
97.9
23
PDB header: protein bindingChain: B: PDB Molecule: toxin protein pare-1;PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
14 c4mctD_
97.5
18
PDB header: toxinChain: D: PDB Molecule: killer protein;PDBTitle: p. vulgaris higba structure, crystal form 1
15 c5ja9D_
97.0
17
PDB header: toxinChain: D: PDB Molecule: toxin higb-2;PDBTitle: crystal structure of the higb2 toxin in complex with nb6
16 c6f8sD_
92.8
21
PDB header: toxinChain: D: PDB Molecule: putative killer protein;PDBTitle: toxin-antitoxin complex grata
17 c2kruA_
77.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: solution nmr structure of the pcp_red domain of light-independent2 protochlorophyllide reductase subunit b from chlorobium tepidum.3 northeast structural genomics consortium target ctr69a
18 c2l09A_
69.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: asr4154 protein;PDBTitle: solution nmr structure of protein asr4154 from nostoc sp. pcc71202 northeast structural genomics consortium target id nsr143
19 c5ifgC_
57.6
17
PDB header: hydrolase/antitoxinChain: C: PDB Molecule: mrna interferase higb;PDBTitle: crystal structure of higa-higb complex from e. coli
20 c4b2gB_
51.8
17
PDB header: signaling proteinChain: B: PDB Molecule: gh3-1 auxin conjugating enzyme;PDBTitle: crystal structure of an indole-3-acetic acid amido synthase from vitis2 vinifera involved in auxin homeostasis
21 c6avhA_
not modelled
51.7
14
PDB header: ligase, plant proteinChain: A: PDB Molecule: gh3.15 acyl acid amido synthetase;PDBTitle: gh3.15 acyl acid amido synthetase
22 c4ewvB_
not modelled
47.3
16
PDB header: ligaseChain: B: PDB Molecule: 4-substituted benzoates-glutamate ligase gh3.12;PDBTitle: crystal structure of gh3.12 in complex with ampcpp
23 c4eplA_
not modelled
46.2
17
PDB header: ligaseChain: A: PDB Molecule: jasmonic acid-amido synthetase jar1;PDBTitle: crystal structure of arabidopsis thaliana gh3.11 (jar1) in complex2 with ja-ile
24 c5kodA_
not modelled
40.0
19
PDB header: ligaseChain: A: PDB Molecule: indole-3-acetic acid-amido synthetase gh3.5;PDBTitle: crystal structure of gh3.5 acyl acid amido synthetase from arabidopsis2 thaliana
25 c2ynmD_
not modelled
15.8
15
PDB header: oxidoreductaseChain: D: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
26 c2lc0A_
not modelled
15.0
9
PDB header: protein bindingChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: rv0020c_nter structure
27 c2apnA_
not modelled
11.8
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein hi1723;PDBTitle: hi1723 solution structure
28 c4le6B_
not modelled
11.8
13
PDB header: hydrolaseChain: B: PDB Molecule: organophosphorus hydrolase;PDBTitle: crystal structure of the phosphotriesterase ophc2 from pseudomonas2 pseudoalcaligenes
29 c4xukB_
not modelled
11.0
19
PDB header: hydrolaseChain: B: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of hydrolase aboph in beta lactamase superfamily
30 d1ugja_
not modelled
8.2
12
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: RIKEN cDNA 2310057j16 protein (KIAA1543)
31 c2lf0A_
not modelled
7.2
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yibl;PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scattering, nesg4 target sfr339/ocsp target sf3636
32 d1p9ea_
not modelled
7.2
17
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Methyl parathion hydrolase
33 c1p9eA_
not modelled
7.2
17
PDB header: hydrolaseChain: A: PDB Molecule: methyl parathion hydrolase;PDBTitle: crystal structure analysis of methyl parathion hydrolase from2 pseudomonas sp wbc-3
34 c1x60A_
not modelled
6.7
18
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
35 c5m5cC_
not modelled
6.6
17
PDB header: transport proteinChain: C: PDB Molecule: calmodulin-regulated spectrin-associated protein 1;PDBTitle: mechanism of microtubule minus-end recognition and protection by2 camsap proteins
36 c2h5gA_
not modelled
5.8
21
PDB header: oxidoreductaseChain: A: PDB Molecule: delta 1-pyrroline-5-carboxylate synthetase;PDBTitle: crystal structure of human pyrroline-5-carboxylate synthetase
37 c6fkfd_
not modelled
5.5
21
PDB header: membrane proteinChain: D: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 1