| 1 |
|
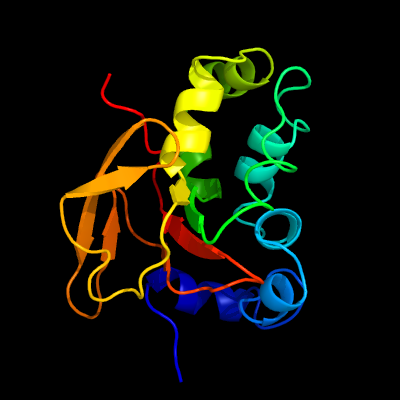
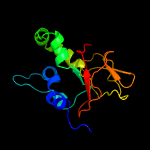
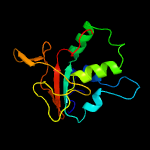
PDB 1nyo chain A
Region: 31 - 193
Aligned: 163
Modelled: 163
Confidence: 100.0%
Identity: 100%
Fold: FAS1 domain
Superfamily: FAS1 domain
Family: FAS1 domain
Phyre2
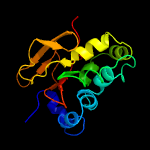
| 2 |
|
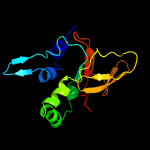
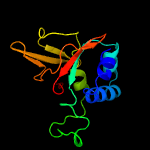
PDB 5nv6 chain A
Region: 50 - 193
Aligned: 139
Modelled: 144
Confidence: 100.0%
Identity: 32%
PDB header:structural protein
Chain: A: PDB Molecule:transforming growth factor-beta-induced protein ig-h3;
PDBTitle: structure of human transforming growth factor beta-induced protein2 (tgfbip).
Phyre2
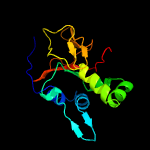
| 3 |
|
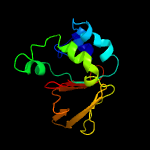
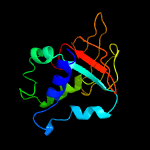
PDB 1x3b chain A
Region: 50 - 193
Aligned: 139
Modelled: 144
Confidence: 100.0%
Identity: 30%
PDB header:cell adhesion
Chain: A: PDB Molecule:transforming growth factor-beta induced protein
PDBTitle: solution structure of the fas1 domain of human transforming2 growth factor-beta induced protein ig-h3
Phyre2
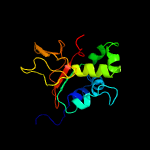
| 4 |
|
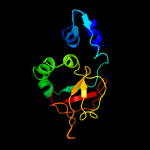
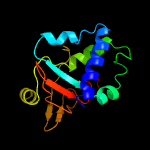
PDB 5yjh chain A
Region: 55 - 191
Aligned: 132
Modelled: 137
Confidence: 100.0%
Identity: 22%
PDB header:cell adhesion
Chain: A: PDB Molecule:periostin;
PDBTitle: structural insights into periostin functions
Phyre2
| 5 |
|
PDB 5wt7 chain A
Region: 57 - 192
Aligned: 131
Modelled: 136
Confidence: 100.0%
Identity: 22%
PDB header:cell adhesion
Chain: A: PDB Molecule:periostin;
PDBTitle: fas1-iv domain of human periostin
Phyre2
| 6 |
|
PDB 1w7e chain A
Region: 59 - 191
Aligned: 127
Modelled: 133
Confidence: 100.0%
Identity: 43%
PDB header:cell adhesion
Chain: A: PDB Molecule:beta-ig-h3/fasciclin;
PDBTitle: nmr ensemble of fasciclin-like protein from rhodobacter sphaeroides
Phyre2
| 7 |
|
PDB 2mxa chain A
Region: 59 - 188
Aligned: 124
Modelled: 130
Confidence: 100.0%
Identity: 40%
PDB header:membrane protein
Chain: A: PDB Molecule:ndh-1 complex sensory subunit cups;
PDBTitle: solution structure of the ndh-1 complex subunit cups from2 thermosynechococcus elongatus
Phyre2
| 8 |
|
PDB 1o70 chain A domain 2
Region: 59 - 193
Aligned: 130
Modelled: 135
Confidence: 100.0%
Identity: 18%
Fold: FAS1 domain
Superfamily: FAS1 domain
Family: FAS1 domain
Phyre2
| 9 |
|
PDB 5n86 chain A
Region: 59 - 192
Aligned: 123
Modelled: 134
Confidence: 100.0%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:stabilin-2;
PDBTitle: crystal structure of fas1 domain of hyaluronic acid receptor stabilin-2 2
Phyre2
| 10 |
|
PDB 1o70 chain A domain 1
Region: 65 - 192
Aligned: 120
Modelled: 128
Confidence: 99.9%
Identity: 23%
Fold: FAS1 domain
Superfamily: FAS1 domain
Family: FAS1 domain
Phyre2
| 11 |
|
PDB 1o70 chain A
Region: 65 - 193
Aligned: 121
Modelled: 129
Confidence: 99.9%
Identity: 26%
PDB header:cell adhesion
Chain: A: PDB Molecule:fasciclin i;
PDBTitle: novel fold revealed by the structure of a fas1 domain pair2 from the insect cell adhesion molecule fasciclin i
Phyre2
| 12 |
|
PDB 2ag3 chain A
Region: 121 - 132
Aligned: 12
Modelled: 12
Confidence: 19.0%
Identity: 42%
PDB header:de novo protein
Chain: A: PDB Molecule:gcn4-pli;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: substitution of the k(15)-l(16) amide with a triazole
Phyre2
| 13 |
|
PDB 1u9f chain A
Region: 121 - 132
Aligned: 12
Modelled: 12
Confidence: 10.8%
Identity: 42%
PDB header:transcription
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)
Phyre2
| 14 |
|
PDB 1u9f chain C
Region: 121 - 132
Aligned: 12
Modelled: 12
Confidence: 10.2%
Identity: 42%
PDB header:transcription
Chain: C: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)
Phyre2
| 15 |
|
PDB 1u9f chain B
Region: 121 - 132
Aligned: 12
Modelled: 12
Confidence: 10.2%
Identity: 42%
PDB header:transcription
Chain: B: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)
Phyre2
| 16 |
|
PDB 1u9f chain D
Region: 121 - 132
Aligned: 12
Modelled: 12
Confidence: 10.0%
Identity: 42%
PDB header:transcription
Chain: D: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)
Phyre2
| 17 |
|
PDB 4v19 chain P
Region: 97 - 109
Aligned: 13
Modelled: 13
Confidence: 7.5%
Identity: 23%
PDB header:ribosome
Chain: P: PDB Molecule:mitoribosomal protein ul15m, mrpl15;
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
Phyre2
| 18 |
|
PDB 2kwr chain A
Region: 179 - 190
Aligned: 12
Modelled: 12
Confidence: 7.5%
Identity: 25%
PDB header:transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution nmr structure of the apoform of nare (nmb1343)
Phyre2