1 c6adqP_
100.0
61
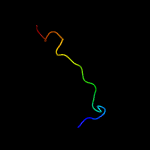
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
2 c2rmzA_
59.7
26
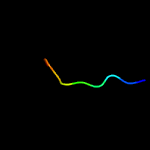
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
3 c5n9yB_
59.1
22
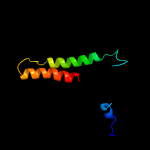
PDB header: membrane proteinChain: B: PDB Molecule: zinc transport protein zntb;PDBTitle: the full-length structure of zntb
4 c4ev6E_
57.6
20
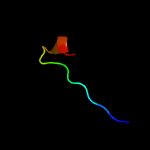
PDB header: metal transportChain: E: PDB Molecule: magnesium transport protein cora;PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
5 c5zazA_
37.0
22
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: solution structure of integrin b2 monomer tranmembrane domain in2 bicelle
6 c2ls2A_
35.4
40
PDB header: metal transportChain: A: PDB Molecule: high affinity copper uptake protein 1;PDBTitle: 1h chemical shift assignments for the first transmembrane domain from2 human copper transport 1
7 c2kncB_
33.9
26
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
8 c2momB_
23.5
24
PDB header: membrane proteinChain: B: PDB Molecule: lysosome-associated membrane glycoprotein 2;PDBTitle: structural insights of tm domain of lamp-2a in dpc micelles
9 c2momC_
23.4
24
PDB header: membrane proteinChain: C: PDB Molecule: lysosome-associated membrane glycoprotein 2;PDBTitle: structural insights of tm domain of lamp-2a in dpc micelles
10 d1eu1a1
15.3
33
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain
11 c5iqjB_
14.5
33
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: 1.9 angstrom crystal structure of protein with unknown function from2 vibrio cholerae.
12 c2d3aJ_
12.2
56
PDB header: ligaseChain: J: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of the maize glutamine synthetase2 complexed with adp and methionine sulfoximine phosphate
13 c4p5nB_
11.5
50
PDB header: structural genomicsChain: B: PDB Molecule: hypothetical protein cnag_02591;PDBTitle: structure of cnag_02591 from cryptococcus neoformans
14 c2w2eA_
10.6
21
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin pip2-7 7;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin, aqy1, in a2 closed conformation at ph 3.5
15 c4baxH_
9.4
50
PDB header: ligaseChain: H: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of glutamine synthetase from streptomyces2 coelicolor
16 c4is4G_
9.2
56
PDB header: ligaseChain: G: PDB Molecule: glutamine synthetase;PDBTitle: the glutamine synthetase from the dicotyledonous plant m. truncatula2 is a decamer
17 c2xczA_
8.6
45
PDB header: immune systemChain: A: PDB Molecule: possible atls1-like light-inducible protein;PDBTitle: crystal structure of macrophage migration inhibitory factor homologue2 from prochlorococcus marinus
18 c3gacD_
8.2
9
PDB header: cytokineChain: D: PDB Molecule: macrophage migration inhibitory factor-likePDBTitle: structure of mif with hpp
19 c6cuqB_
8.2
27
PDB header: cytokineChain: B: PDB Molecule: macrophage migration inhibitory factor-like protein;PDBTitle: crystal structure of macrophage migration inhibitory factor-like2 protein (ehmif) from entamoeba histolytica
20 c4c84B_
7.8
27
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase znrf3;PDBTitle: zebrafish znrf3 ectodomain crystal form i
21 c3ug9A_
not modelled
7.2
9
PDB header: membrane proteinChain: A: PDB Molecule: archaeal-type opsin 1, archaeal-type opsin 2;PDBTitle: crystal structure of the closed state of channelrhodopsin
22 d1uiza_
not modelled
7.1
27
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related
23 c3njaC_
not modelled
6.5
22
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: probable ggdef family protein;PDBTitle: the crystal structure of the pas domain of a ggdef family protein from2 chromobacterium violaceum atcc 12472.
24 c3icfB_
not modelled
6.2
26
PDB header: hydrolaseChain: B: PDB Molecule: serine/threonine-protein phosphatase t;PDBTitle: structure of protein serine/threonine phosphatase from saccharomyces2 cerevisiae with similarity to human phosphatase pp5
25 d1pq1a_
not modelled
6.1
19
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death
26 c2mkvA_
not modelled
5.9
21
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
27 c1hznA_
not modelled
5.9
27
PDB header: hormone/growth factorChain: A: PDB Molecule: cholecystokinin type a receptor;PDBTitle: nmr solution structure of the third extracellular loop of2 the cholecystokinin a receptor
28 c5unqF_
not modelled
5.9
23
PDB header: hydrolaseChain: F: PDB Molecule: putative tautomerase;PDBTitle: crystal structure of pt0534 inactivated by 2-oxo-3-pentynoate
29 c2mj2A_
not modelled
5.8
25
PDB header: viral proteinChain: A: PDB Molecule: agnoprotein;PDBTitle: structure of the dimerization domain of the human polyoma, jc virus2 agnoprotein is an amphipathic alpha-helix.
30 c2os5C_
not modelled
5.8
9
PDB header: cytokineChain: C: PDB Molecule: acemif;PDBTitle: macrophage migration inhibitory factor from ancylostoma ceylanicum
31 c3rhtB_
not modelled
5.7
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: (gatase1)-like protein;PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
32 d1dpta_
not modelled
5.6
18
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related
33 c2jo1A_
not modelled
5.5
38
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
34 c2jp3A_
not modelled
5.5
25
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
35 c6cfwI_
not modelled
5.4
25
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
36 c2m59B_
not modelled
5.3
13
PDB header: transferaseChain: B: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
37 c2m59A_
not modelled
5.3
13
PDB header: transferaseChain: A: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
38 c2ks1A_
not modelled
5.3
17
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
39 c2jwaA_
not modelled
5.3
17
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
40 c4dh4A_
not modelled
5.2
9
PDB header: isomeraseChain: A: PDB Molecule: mif;PDBTitle: macrophage migration inhibitory factor toxoplasma gondii
41 c3mk7F_
not modelled
5.2
16
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
42 d1hfoa_
not modelled
5.2
18
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: MIF-related