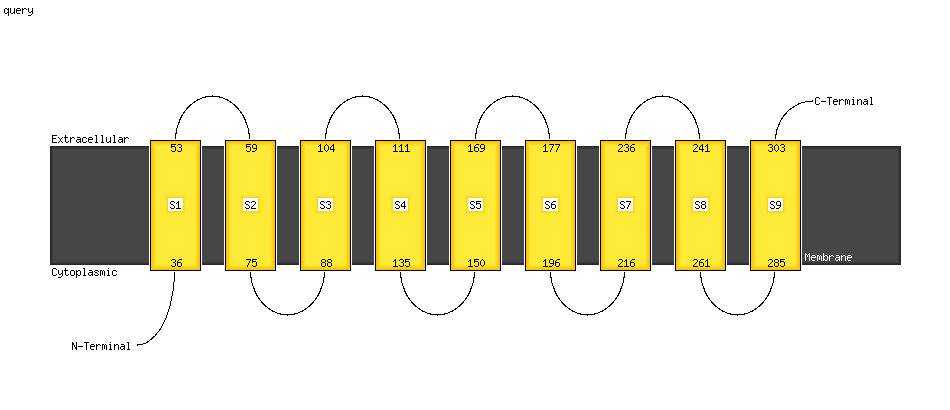
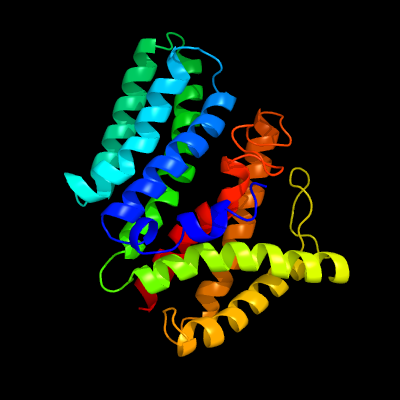
| 1 | c4q2eA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidate cytidylyltransferase;
PDBTitle: crystal structure of an intramembrane cdp-dag synthetase central for2 phospholipid biosynthesis (s200c/s258c, active mutant)
|
|
|
|
| 2 | c4q2gA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidate cytidylyltransferase;
PDBTitle: crystal structure of an intramembrane cdp-dag synthetase central for2 phospholipid biosynthesis (s200c/s223c, inactive mutant)
|
|
|
|
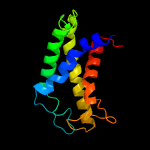
| 3 | c5gufA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:cdp-archaeol synthase;
PDBTitle: structural insight into an intramembrane enzyme for archaeal membrane2 lipids biosynthesis
|
|
|
|
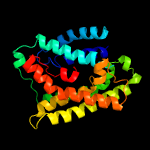
| 4 | c3b9yA_
|
|
 |
96.0 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
|
|
|
|
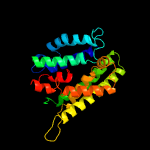
| 5 | c5aexJ_
|
|
 |
88.2 |
12 |
PDB header:membrane protein
Chain: J: PDB Molecule:ammonium transporter mep2;
PDBTitle: crystal structure of saccharomyces cerevisiae mep2
|
|
|
|
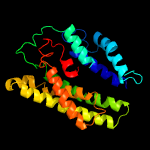
| 6 | c5aexB_
|
|
 |
85.3 |
17 |
PDB header:membrane protein
Chain: B: PDB Molecule:ammonium transporter mep2;
PDBTitle: crystal structure of saccharomyces cerevisiae mep2
|
|
|
|
| 7 | c2nuuF_
|
|
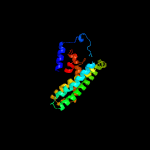 |
83.0 |
14 |
PDB header:transport protein/signaling protein
Chain: F: PDB Molecule:ammonia channel;
PDBTitle: regulating the escherichia coli ammonia channel: the crystal structure2 of the amtb-glnk complex
|
|
|
|
| 8 | c6eu6A_
|
|
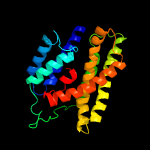 |
77.0 |
19 |
PDB header:membrane protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: sensor amt protein
|
|
|
|
| 9 | c3hd6A_
|
|
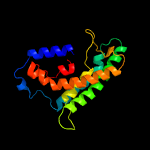 |
75.7 |
12 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
|
|
|
|
| 10 | c5aezA_
|
|
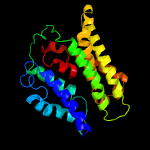 |
62.7 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:mep2;
PDBTitle: crystal structure of candida albicans mep2
|
|
|
|
| 11 | c3r2uC_
|
|
 |
61.7 |
22 |
PDB header:hydrolase
Chain: C: PDB Molecule:metallo-beta-lactamase family protein;
PDBTitle: 2.1 angstrom resolution crystal structure of metallo-beta-lactamase2 from staphylococcus aureus subsp. aureus col
|
|
|
|
| 12 | d2p97a1
|
|
 |
57.4 |
30 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Ava3068-like |
|
|
|
| 13 | c2gcuD_
|
|
 |
57.1 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative hydroxyacylglutathione hydrolase 3;
PDBTitle: x-ray structure of gene product from arabidopsis thaliana at1g53580
|
|
|
|
| 14 | c4chlA_
|
|
 |
55.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:persulfide dioxygenase ethe1, mitochondrial;
PDBTitle: human ethylmalonic encephalopathy protein 1 (hethe1)
|
|
|
|
| 15 | c2xf4A_
|
|
 |
54.3 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydroxyacylglutathione hydrolase;
PDBTitle: crystal structure of salmonella enterica serovar2 typhimurium ycbl
|
|
|
|
| 16 | c5ve5C_
|
|
 |
54.1 |
12 |
PDB header:oxidoreductase, transferase
Chain: C: PDB Molecule:bpprf;
PDBTitle: crystal structure of persulfide dioxygenase rhodanese fusion protein2 with rhodanese domain inactivating mutation (c314s) from burkholderia3 phytofirmans in complex with glutathione
|
|
|
|
| 17 | c4yskA_
|
|
 |
53.2 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-lactamase domain protein;
PDBTitle: crystal structure of apo-form sdoa from pseudomonas putida
|
|
|
|
| 18 | c2mc5A_
|
|
 |
51.8 |
62 |
PDB header:protein binding
Chain: A: PDB Molecule:45l;
PDBTitle: a bacteriophage transcription regulator inhibits bacterial2 transcription initiation by -factor displacement
|
|
|
|
| 19 | c3tp9B_
|
|
 |
50.8 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactamase and rhodanese domain protein;
PDBTitle: crystal structure of alicyclobacillus acidocaldarius protein with2 beta-lactamase and rhodanese domains
|
|
|
|
| 20 | c4ysbB_
|
|
 |
50.6 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallo-beta-lactamase family protein;
PDBTitle: crystal structure of ethe1 from myxococcus xanthus
|
|
|
|
| 21 | c4efzB_ |
|
not modelled |
50.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallo-beta-lactamase family protein;
PDBTitle: crystal structure of a hypothetical metallo-beta-lactamase from2 burkholderia pseudomallei
|
|
|
| 22 | c2kogA_ |
|
not modelled |
47.4 |
30 |
PDB header:membrane protein
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: lipid-bound synaptobrevin solution nmr structure
|
|
|
| 23 | d1mqoa_ |
|
not modelled |
45.8 |
19 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 24 | d1qh5a_ |
|
not modelled |
43.1 |
11 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Glyoxalase II (hydroxyacylglutathione hydrolase) |
|
|
| 25 | c4ad9E_ |
|
not modelled |
41.9 |
32 |
PDB header:hydrolase
Chain: E: PDB Molecule:beta-lactamase-like protein 2;
PDBTitle: crystal structure of human lactb2.
|
|
|
| 26 | c4awyB_ |
|
not modelled |
41.9 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallo-beta-lactamase aim-1;
PDBTitle: crystal structure of the mobile metallo-beta-lactamase aim-1 from2 pseudomonas aeruginosa: insights into antibiotic binding and the role3 of gln157
|
|
|
| 27 | d1xm8a_ |
|
not modelled |
40.6 |
13 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Glyoxalase II (hydroxyacylglutathione hydrolase) |
|
|
| 28 | c5sxpG_ |
|
not modelled |
40.4 |
20 |
PDB header:signaling protein/ligase
Chain: G: PDB Molecule:e3 ubiquitin-protein ligase itchy homolog;
PDBTitle: structural basis for the interaction between itch prr and beta-pix
|
|
|
| 29 | d1u7ga_ |
|
not modelled |
37.4 |
18 |
Fold:Ammonium transporter
Superfamily:Ammonium transporter
Family:Ammonium transporter |
|
|
| 30 | d2aioa1 |
|
not modelled |
34.8 |
21 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 31 | d1znba_ |
|
not modelled |
34.8 |
19 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 32 | d1bh9b_ |
|
not modelled |
29.0 |
27 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
|
|
| 33 | c5i0pB_ |
|
not modelled |
28.8 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactamase domain protein;
PDBTitle: crystal structure of a beta-lactamase domain protein from burkholderia2 ambifaria
|
|
|
| 34 | c6h0cA_ |
|
not modelled |
28.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: flv1 flavodiiron core from synechocystis sp. pcc6803
|
|
|
| 35 | c3hnnD_ |
|
not modelled |
28.5 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative diflavin flavoprotein a 5;
PDBTitle: crystal structure of putative diflavin flavoprotein a 5 (fragment 1-2 254) from nostoc sp. pcc 7120, northeast structural genomics3 consortium target nsr435a
|
|
|
| 36 | d2qeda1 |
|
not modelled |
28.2 |
10 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Glyoxalase II (hydroxyacylglutathione hydrolase) |
|
|
| 37 | c3c1iA_ |
|
not modelled |
27.5 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonia channel;
PDBTitle: substrate binding, deprotonation and selectivity at the periplasmic2 entrance of the e. coli ammonia channel amtb
|
|
|
| 38 | c2p18A_ |
|
not modelled |
26.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:glyoxalase ii;
PDBTitle: crystal structure of the leishmania infantum glyoxalase ii
|
|
|
| 39 | c2zo4A_ |
|
not modelled |
25.4 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:metallo-beta-lactamase family protein;
PDBTitle: crystal structure of metallo-beta-lactamase family protein ttha14292 from thermus thermophilus hb8
|
|
|
| 40 | c1vmeB_ |
|
not modelled |
23.8 |
17 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoprotein;
PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
|
|
|
| 41 | c5ja9D_ |
|
not modelled |
23.1 |
42 |
PDB header:toxin
Chain: D: PDB Molecule:toxin higb-2;
PDBTitle: crystal structure of the higb2 toxin in complex with nb6
|
|
|
| 42 | c2zwrA_ |
|
not modelled |
22.0 |
42 |
PDB header:hydrolase
Chain: A: PDB Molecule:metallo-beta-lactamase superfamily protein;
PDBTitle: crystal structure of ttha1623 from thermus thermophilus hb8
|
|
|
| 43 | c4v0hC_ |
|
not modelled |
21.4 |
41 |
PDB header:hydrolase
Chain: C: PDB Molecule:metallo-beta-lactamase domain-containing protein 1 1;
PDBTitle: human metallo beta lactamase domain containing protein 1 (hmblac1)
|
|
|
| 44 | d1m2xa_ |
|
not modelled |
21.2 |
17 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 45 | d1ycga2 |
|
not modelled |
20.8 |
15 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:ROO N-terminal domain-like |
|
|
| 46 | c4wd6B_ |
|
not modelled |
20.2 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallo-beta-lactamase;
PDBTitle: crystal structure of dim-1 metallo-beta-lactamase
|
|
|
| 47 | c6n36A_ |
|
not modelled |
20.1 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:beta-lactamase;
PDBTitle: beta-lactamase from chitinophaga pinensis
|
|
|
| 48 | c3dh4A_ |
|
not modelled |
20.0 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
|
|
|
| 49 | c3l6nA_ |
|
not modelled |
19.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:metallo-beta-lactamase;
PDBTitle: crystal structure of metallo-beta-lactamase ind-7
|
|
|
| 50 | c5al6A_ |
|
not modelled |
19.4 |
25 |
PDB header:structural protein
Chain: A: PDB Molecule:anastral spindle 2;
PDBTitle: central coiled-coil domain (cccd) of drosophila melanogaster ana2. a2 natural, parallel, tetrameric coiled-coil bundle.
|
|
|
| 51 | c2jpjA_ |
|
not modelled |
19.1 |
40 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit alpha;
PDBTitle: lactococcin g-a in dpc
|
|
|
| 52 | c2jplA_ |
|
not modelled |
19.1 |
40 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit alpha;
PDBTitle: lactococcin g-a in tfe
|
|
|
| 53 | c2q9uB_ |
|
not modelled |
18.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:a-type flavoprotein;
PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
|
|
|
| 54 | c6oitG_ |
|
not modelled |
18.5 |
25 |
PDB header:plant protein
Chain: G: PDB Molecule:protein chromatin remodeling 35;
PDBTitle: cryoem structure of arabidopsis ddr' complex (drd1 peptide-dms3-rdm1)
|
|
|
| 55 | c6aufB_ |
|
not modelled |
18.0 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactamase-like protein;
PDBTitle: crystal structure of metalo beta lactamases mim-1 from novosphingobium2 pentaromativorans
|
|
|
| 56 | c3lvzA_ |
|
not modelled |
17.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:blr6230 protein;
PDBTitle: new refinement of the crystal structure of bjp-1, a subclass b32 metallo-beta-lactamase of bradyrhizobium japonicum
|
|
|
| 57 | c2yz3B_ |
|
not modelled |
17.1 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:metallo-beta-lactamase;
PDBTitle: crystallographic investigation of inhibition mode of the2 vim-2 metallo-beta-lactamase from pseudomonas aeruginosa3 with mercaptocarboxylate inhibitor
|
|
|
| 58 | c5k0wA_ |
|
not modelled |
17.0 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:class b carbapenemase gob-18;
PDBTitle: crystal structure of the metallo-beta-lactamase gob-18 from2 elizabethkingia meningoseptica
|
|
|
| 59 | c6qrqB_ |
|
not modelled |
17.0 |
18 |
PDB header:signaling protein
Chain: B: PDB Molecule:oxygen-binding diiron protein;
PDBTitle: apo conformation of chemotaxis sensor odp
|
|
|
| 60 | d1nxca_ |
|
not modelled |
16.9 |
13 |
Fold:alpha/alpha toroid
Superfamily:Seven-hairpin glycosidases
Family:Class I alpha-1;2-mannosidase, catalytic domain |
|
|
| 61 | d1dl2a_ |
|
not modelled |
16.8 |
14 |
Fold:alpha/alpha toroid
Superfamily:Seven-hairpin glycosidases
Family:Class I alpha-1;2-mannosidase, catalytic domain |
|
|
| 62 | c2kdcC_ |
|
not modelled |
16.3 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: nmr solution structure of e. coli diacylglycerol kinase2 (dagk) in dpc micelles
|
|
|
| 63 | c5aebA_ |
|
not modelled |
16.1 |
4 |
PDB header:hydrolase
Chain: A: PDB Molecule:lra-12;
PDBTitle: crystal structure of the class b3 di-zinc metallo-beta-lactamase lra-2 12 from an alaskan soil metagenome.
|
|
|
| 64 | c2e55D_ |
|
not modelled |
16.0 |
43 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structure of aq2163 protein from aquifex aeolicus
|
|
|
| 65 | c3bboU_ |
|
not modelled |
15.6 |
22 |
PDB header:ribosome
Chain: U: PDB Molecule:ribosomal protein l22;
PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
|
|
|
| 66 | d2gmna1 |
|
not modelled |
15.6 |
26 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 67 | d2ey4a2 |
|
not modelled |
15.3 |
35 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
|
|
| 68 | c6ixhQ_ |
|
not modelled |
15.3 |
17 |
PDB header:membrane protein
Chain: Q: PDB Molecule:type vi secretion system tssm;
PDBTitle: type vi secretion system membrane core complex
|
|
|
| 69 | c4binA_ |
|
not modelled |
15.1 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase amic;
PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
|
|
|
| 70 | d2nn6h2 |
|
not modelled |
14.9 |
33 |
Fold:Barrel-sandwich hybrid
Superfamily:Ribosomal L27 protein-like
Family:ECR1 N-terminal domain-like |
|
|
| 71 | c1s1iN_ |
|
not modelled |
14.7 |
33 |
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l17-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
|
|
|
| 72 | d2gycq1 |
|
not modelled |
14.1 |
29 |
Fold:Ribosomal protein L22
Superfamily:Ribosomal protein L22
Family:Ribosomal protein L22 |
|
|
| 73 | d1ko3a_ |
|
not modelled |
14.1 |
19 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Zn metallo-beta-lactamase |
|
|
| 74 | c3pisA_ |
|
not modelled |
13.5 |
63 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:kazal-type serine protease inhibitor spi-1;
PDBTitle: crystal structure of carcinoscorpius rotundicauda serine protease2 inhibitor domain 1
|
|
|
| 75 | c3dmpD_ |
|
not modelled |
13.4 |
38 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: 2.6 a crystal structure of uracil phosphoribosyltransferase from2 burkholderia pseudomallei
|
|
|
| 76 | c6dn4A_ |
|
not modelled |
13.4 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:beta-lactamase;
PDBTitle: cronobacter sakazakii (enterobacter sakazakii) metallo-beta-lactamse2 harldq motif
|
|
|
| 77 | c5j72B_ |
|
not modelled |
12.9 |
100 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative n-acetylmuramoyl-l-alanine amidase,autolysin cwp6;
PDBTitle: cwp6 from clostridium difficile
|
|
|
| 78 | c3sd9B_ |
|
not modelled |
12.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-lactamase;
PDBTitle: crystal structure of serratia fonticola sfh-i: source of the2 nucleophile in the catalytic mechanism of mono-zinc metallo-beta-3 lactamases
|
|
|
| 79 | c3u5eP_ |
|
not modelled |
12.8 |
33 |
PDB header:ribosome
Chain: P: PDB Molecule:60s ribosomal protein l17-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 60s subunit, ribosome a
|
|
|
| 80 | d1jv1a_ |
|
not modelled |
12.4 |
42 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 81 | d1i5ea_ |
|
not modelled |
12.4 |
43 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 82 | c1ychD_ |
|
not modelled |
12.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitric oxide reductase;
PDBTitle: x-ray crystal structures of moorella thermoacetica fpra. novel diiron2 site structure and mechanistic insights into a scavenging nitric3 oxide reductase
|
|
|
| 83 | d2icya2 |
|
not modelled |
12.3 |
50 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
|
|
| 84 | c3jywN_ |
|
not modelled |
11.9 |
33 |
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l17(a);
PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
|
|
|
| 85 | d1k8wa5 |
|
not modelled |
11.9 |
35 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
|
|
| 86 | c3zf7R_ |
|
not modelled |
11.8 |
22 |
PDB header:ribosome
Chain: R: PDB Molecule:60s ribosomal protein l17, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 87 | c1k8wA_ |
|
not modelled |
11.2 |
38 |
PDB header:lyase/rna
Chain: A: PDB Molecule:trna pseudouridine synthase b;
PDBTitle: crystal structure of the e. coli pseudouridine synthase2 trub bound to a t stem-loop rna
|
|
|
| 88 | c5e38D_ |
|
not modelled |
11.2 |
33 |
PDB header:transferase
Chain: D: PDB Molecule:uracil phosphoribosyltransferase;
PDBTitle: structural basis of mapping the spontaneous mutations with 5-2 flourouracil in uracil phosphoribosyltransferase from mycobacterium3 tuberculosis
|
|
|
| 89 | c3oc9A_ |
|
not modelled |
11.2 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine pyrophosphorylase;
PDBTitle: crystal structure of putative udp-n-acetylglucosamine2 pyrophosphorylase from entamoeba histolytica
|
|
|
| 90 | c1e5dA_ |
|
not modelled |
11.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin\:oxygen oxidoreductase;
PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe desulfovibrio2 gigas
|
|
|
| 91 | c2fhxB_ |
|
not modelled |
10.9 |
28 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:spm-1;
PDBTitle: pseudomonas aeruginosa spm-1 metallo-beta-lactamase
|
|
|
| 92 | d1sgva2 |
|
not modelled |
10.8 |
30 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
|
|
| 93 | c3zf7Y_ |
|
not modelled |
10.8 |
44 |
PDB header:ribosome
Chain: Y: PDB Molecule:60s ribosomal protein l24, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 94 | c3pesA_ |
|
not modelled |
10.8 |
67 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein gp49;
PDBTitle: crystal structure of uncharacterized protein from pseudomonas phage2 yua
|
|
|
| 95 | c6cqsA_ |
|
not modelled |
10.8 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-lactamase;
PDBTitle: sediminispirochaeta smaragdinae sps-1 metallo-beta-lactamase
|
|
|
| 96 | c2zvrA_ |
|
not modelled |
10.8 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related protein from2 thermotoga maritima
|
|
|
| 97 | c6nu9A_ |
|
not modelled |
10.7 |
33 |
PDB header:viral protein
Chain: A: PDB Molecule:zinc-binding non-structural protein;
PDBTitle: crystal structure of a zinc-binding non-structural protein from the2 hepatitis e virus
|
|
|
| 98 | c3vqzA_ |
|
not modelled |
10.7 |
7 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:metallo-beta-lactamase;
PDBTitle: crystal structure of metallo-beta-lactamase, smb-1, in a complex with2 mercaptoacetic acid
|
|
|
| 99 | c3fvyA_ |
|
not modelled |
10.6 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidyl-peptidase 3;
PDBTitle: crystal structure of human dipeptidyl peptidase iii
|
|
|