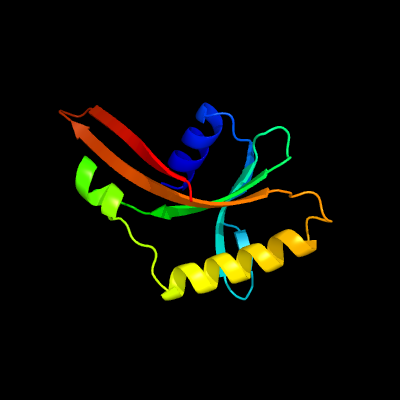
1 c3fovA_
100.0
41
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0102 protein rpa0323;PDBTitle: crystal structure of protein rpa0323 of unknown function from2 rhodopseudomonas palustris
2 d1gefa_
97.4
22
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like
3 d1ob8a_
96.9
24
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like
4 c2eo0A_
96.6
22
PDB header: dna binding proteinChain: A: PDB Molecule: hypothetical protein st1444;PDBTitle: crystal structure of holliday junction resolvase st1444
5 c2wj0B_
96.2
23
PDB header: hydrolase/dnaChain: B: PDB Molecule: archaeal hjc;PDBTitle: crystal structures of holliday junction resolvases from2 archaeoglobus fulgidus bound to dna substrate
6 c5gkeB_
95.3
26
PDB header: hydrolase/dnaChain: B: PDB Molecule: endonuclease endoms;PDBTitle: structure of endoms-dsdna1 complex
7 d1hh1a_
95.3
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like
8 c2vldA_
94.3
29
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease nucs;PDBTitle: crystal structure of a repair endonuclease from pyrococcus abyssi
9 c4dapA_
94.0
23
PDB header: dna binding proteinChain: A: PDB Molecule: sugar fermentation stimulation protein a;PDBTitle: the structure of escherichia coli sfsa
10 c3dnxA_
93.7
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein spo1766;PDBTitle: spo1766 protein of unknown function from silicibacter pomeroyi.
11 d1y88a2
93.7
22
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: MRR-like
12 c1y88A_
93.6
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein af1548;PDBTitle: crystal structure of protein of unknown function af1548
13 c4davA_
91.2
22
PDB header: hydrolase/dnaChain: A: PDB Molecule: sugar fermentation stimulation protein homolog;PDBTitle: the structure of pyrococcus furiosus sfsa in complex with dna
14 d2inba1
83.8
25
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: XisH-like
15 d2okfa1
81.9
23
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: XisH-like
16 c4qbnA_
79.9
19
PDB header: hydrolaseChain: A: PDB Molecule: nuclease;PDBTitle: vrr_nuc domain
17 d1xmxa_
78.4
27
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hypothetical protein VC1899
18 d1cw0a_
73.9
25
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Very short patch repair (VSR) endonuclease
19 c4oc8A_
73.9
28
PDB header: hydrolaseChain: A: PDB Molecule: restriction endonuclease aspbhi;PDBTitle: dna modification-dependent restriction endonuclease aspbhi
20 d1m0da_
70.2
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Endonuclease I (Holliday junction resolvase)
21 c4r8aF_
not modelled
68.5
29
PDB header: hydrolase/dnaChain: F: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of pafan1 - 5' flap dna complex
22 c4ribA_
not modelled
63.7
30
PDB header: hydrolase/dnaChain: A: PDB Molecule: fanconi-associated nuclease 1;PDBTitle: fan1 nuclease bound to 5' phosphorylated p(dt) single flap dna
23 c4qboA_
not modelled
58.0
21
PDB header: hydrolaseChain: A: PDB Molecule: nuclease;PDBTitle: vrr_nuc domain
24 c2ixsB_
not modelled
47.5
18
PDB header: hydrolaseChain: B: PDB Molecule: sdai restriction endonuclease;PDBTitle: structure of sdai restriction endonuclease
25 c4xqkB_
not modelled
45.4
25
PDB header: hydrolase/dnaChain: B: PDB Molecule: llabiii;PDBTitle: atp-dependent type isp restriction-modification enzyme llabiii bound2 to dna
26 d1xdfa1
not modelled
42.5
12
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like
27 c4onbA_
not modelled
26.6
15
PDB header: hydrolaseChain: A: PDB Molecule: crispr-associated exonuclease, cas4 family;PDBTitle: crystal structure of crispr-associated exonuclease (cas4 family) from2 pyrobaculum calidifontis jcm 11548
28 c2k7hA_
not modelled
25.5
10
PDB header: allergenChain: A: PDB Molecule: stress-induced protein sam22;PDBTitle: nmr solution structure of soybean allergen gly m 4
29 d2o8ra3
not modelled
25.2
44
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain
30 c3bt3B_
not modelled
22.9
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: glyoxalase-related enzyme, arac type;PDBTitle: crystal structure of a glyoxalase-related enzyme from clostridium2 phytofermentans
31 c5jhfA_
not modelled
21.9
30
PDB header: protein transportChain: A: PDB Molecule: klth0d11660p;PDBTitle: crystal structure of atg13(17br)-atg13(17lr)-atg17-atg29-atg31 complex
32 c4g6vE_
not modelled
21.7
19
PDB header: toxinChain: E: PDB Molecule: adhesin/hemolysin;PDBTitle: cdia-ct/cdii toxin and immunity complex from burkholderia pseudomallei
33 c2o8rA_
not modelled
18.2
44
PDB header: transferaseChain: A: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
34 d2j9ga3
not modelled
17.3
14
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
35 d1puza_
not modelled
17.3
55
Fold: YgfY-likeSuperfamily: YgfY-likeFamily: YgfY-like
36 d1vsra_
not modelled
17.1
26
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Very short patch repair (VSR) endonuclease
37 d1icxa_
not modelled
16.9
12
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like
38 c5jvvA_
not modelled
16.8
67
PDB header: transferaseChain: A: PDB Molecule: beta-1,3-glucosyltransferase;PDBTitle: crystal structure and characterization an elongating gh family 162 beta-1,3-glucosyltransferase
39 c1x6iB_
not modelled
16.5
64
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ygfy;PDBTitle: crystal structure of ygfy from escherichia coli
40 c2qv6D_
not modelled
15.9
11
PDB header: hydrolaseChain: D: PDB Molecule: gtp cyclohydrolase iii;PDBTitle: gtp cyclohydrolase iii from m. jannaschii (mj0145)2 complexed with gtp and metal ions
41 c2jr5A_
not modelled
15.3
73
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0350 protein vc_2471;PDBTitle: solution structure of upf0350 protein vc_2471. northeast structural2 genomics target vcr36
42 c1gndA_
not modelled
14.5
15
PDB header: gtpase activationChain: A: PDB Molecule: guanine nucleotide dissociation inhibitor;PDBTitle: guanine nucleotide dissociation inhibitor, alpha-isoform
43 d1iowa2
not modelled
13.9
26
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: ATP-binding domain of peptide synthetases
44 d2i7ra1
not modelled
12.8
16
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Antibiotic resistance proteins
45 d2fiwa1
not modelled
12.8
26
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
46 d2jfga1
not modelled
12.2
30
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
47 c3g12A_
not modelled
12.1
15
PDB header: lyaseChain: A: PDB Molecule: putative lactoylglutathione lyase;PDBTitle: crystal structure of a putative lactoylglutathione lyase from2 bdellovibrio bacteriovorus
48 d1miua5
not modelled
12.0
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
49 d1y0ka1
not modelled
12.0
54
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: PA4535-like
50 d1w36b3
not modelled
12.0
15
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Exodeoxyribonuclease V beta chain (RecB), C-terminal domain
51 c3bpqD_
not modelled
11.8
25
PDB header: toxinChain: D: PDB Molecule: toxin rele3;PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
52 d1kjqa3
not modelled
11.4
17
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
53 d1e4ea2
not modelled
11.3
21
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: ATP-binding domain of peptide synthetases
54 c4ic1D_
not modelled
11.3
27
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of sso0001
55 c3g3oA_
not modelled
11.1
36
PDB header: biosynthetic proteinChain: A: PDB Molecule: vacuolar transporter chaperone 2;PDBTitle: crystal structure of the cytoplasmic tunnel domain in yeast2 vtc2p
56 c2duwA_
not modelled
10.7
15
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
57 c2cl2A_
not modelled
10.4
67
PDB header: hydrolaseChain: A: PDB Molecule: putative laminarinase;PDBTitle: endo-1,3(4)-beta-glucanase from phanerochaete chrysosporium, solved2 using native sulfur sad, exhibiting intact heptasaccharide3 glycosylation
58 c4hpqA_
not modelled
10.2
30
PDB header: protein transportChain: A: PDB Molecule: atg29;PDBTitle: crystal structure of the atg17-atg31-atg29 complex
59 c4hpqD_
not modelled
10.2
30
PDB header: protein transportChain: D: PDB Molecule: atg29;PDBTitle: crystal structure of the atg17-atg31-atg29 complex
60 d2ifaa1
not modelled
9.8
17
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
61 c3p1wA_
not modelled
9.6
28
PDB header: protein transportChain: A: PDB Molecule: rabgdi protein;PDBTitle: crystal structure of rab gdi from plasmodium falciparum, pfl2060c
62 c3wdvB_
not modelled
9.6
50
PDB header: hydrolaseChain: B: PDB Molecule: beta-1,3-1,4-glucanase;PDBTitle: the complex structure of ptlic16a with cellotetraose
63 c3r23B_
not modelled
9.6
22
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: crystal structure of d-alanine--d-alanine ligase from bacillus2 anthracis
64 c2mx0A_
not modelled
9.2
26
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein hp_0268;PDBTitle: solution structure of hp0268 from helicobacter pylori
65 c1z8rA_
not modelled
8.9
25
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
66 c3kwkA_
not modelled
8.8
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh dehydrogenase/nad(p)h nitroreductase;PDBTitle: crystal structure of putative nadh dehydrogenase/nad(p)h2 nitroreductase (np_809094.1) from bacteroides thetaiotaomicron vpi-3 5482 at 1.54 a resolution
67 d1j6ua1
not modelled
8.8
19
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
68 d1iyjb5
not modelled
8.6
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
69 c2pvpB_
not modelled
8.5
10
PDB header: ligaseChain: B: PDB Molecule: d-alanine-d-alanine ligase;PDBTitle: crystal structure of d-alanine-d-alanine ligase from helicobacter2 pylori
70 c2m5tA_
not modelled
8.2
25
PDB header: viral proteinChain: A: PDB Molecule: human rhinovirus 2a proteinase;PDBTitle: solution structure of the 2a proteinase from a common cold agent,2 human rhinovirus rv-c02, strain w12
71 c4eo3A_
not modelled
8.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein/nadh dehydrogenase;PDBTitle: peroxiredoxin nitroreductase fusion enzyme
72 d2ayxa2
not modelled
8.0
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: RcsC linker domain-like
73 c3r6aB_
not modelled
7.9
15
PDB header: isomerase, lyaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein (hypothetical protein2 mm_3218) from methanosarcina mazei.
74 c6qb7A_
not modelled
7.8
28
PDB header: signaling proteinChain: A: PDB Molecule: btb/poz domain-containing protein kctd16;PDBTitle: structure of the h1 domain of human kctd16
75 c3ey5A_
not modelled
7.7
5
PDB header: transferaseChain: A: PDB Molecule: acetyltransferase-like, gnat family;PDBTitle: putative acetyltransferase from gnat family from bacteroides2 thetaiotaomicron.
76 d2hrva_
not modelled
7.7
17
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold
77 d3etja3
not modelled
7.5
6
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: BC ATP-binding domain-like
78 d1ifva_
not modelled
7.4
9
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like
79 c3h1tA_
not modelled
7.3
18
PDB header: hydrolaseChain: A: PDB Molecule: type i site-specific restriction-modificationPDBTitle: the fragment structure of a putative hsdr subunit of a type2 i restriction enzyme from vibrio vulnificus yj016
80 d1cjxa1
not modelled
7.2
13
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases
81 c2iyeC_
not modelled
7.2
28
PDB header: hydrolaseChain: C: PDB Molecule: copper-transporting atpase;PDBTitle: structure of catalytic cpx-atpase domain copb-b
82 d1fm4a_
not modelled
7.1
15
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like
83 c3cdlA_
not modelled
7.1
14
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator aefr;PDBTitle: crystal structure of a tetr family transcriptional regulator from2 pseudomonas syringae pv. tomato str. dc3000
84 d1kqba_
not modelled
7.0
9
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
85 c3g5oC_
not modelled
7.0
20
PDB header: toxin/antitoxinChain: C: PDB Molecule: uncharacterized protein rv2866;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
86 c6gq9A_
not modelled
6.8
11
PDB header: allergenChain: A: PDB Molecule: major allergen cor a 1.0401;PDBTitle: solution structure of the hazel allergen cor a 1.0401
87 d1jg5a_
not modelled
6.8
31
Fold: GTP cyclohydrolase I feedback regulatory protein, GFRPSuperfamily: GTP cyclohydrolase I feedback regulatory protein, GFRPFamily: GTP cyclohydrolase I feedback regulatory protein, GFRP
88 c1wv9B_
not modelled
6.8
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: rhodanese homolog tt1651;PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
89 c4g6xA_
not modelled
6.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: crystal structure of glyoxalase/bleomycin resistance protein from2 catenulispora acidiphila.
90 d1qmra_
not modelled
6.7
13
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like
91 c3r3pB_
not modelled
6.7
10
PDB header: hydrolaseChain: B: PDB Molecule: mobile intron protein;PDBTitle: homing endonuclease i-bth0305i catalytic domain
92 d1p3da1
not modelled
6.7
24
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
93 d1o1xa_
not modelled
6.6
19
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB
94 c3zi1A_
not modelled
6.6
21
PDB header: isomeraseChain: A: PDB Molecule: glyoxalase domain-containing protein 4;PDBTitle: crystal structure of human glyoxalase domain-containing protein 42 (glod4)
95 c2rk9B_
not modelled
6.6
2
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: the crystal structure of a glyoxalase/bleomycin resistance2 protein/dioxygenase superfamily member from vibrio splendidus 12b01
96 d1p9oa_
not modelled
6.4
20
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like
97 c4kh8A_
not modelled
6.3
40
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a lipocalin-like protein (ef0376) from2 enterococcus faecalis v583 at 1.60 a resolution
98 c3of4A_
not modelled
6.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a fmn/fad- and nad(p)h-dependent nitroreductase2 (nfnb, il2077) from idiomarina loihiensis l2tr at 1.90 a resolution
99 c2mxqA_
not modelled
6.2
67
PDB header: antimicrobial proteinChain: A: PDB Molecule: paneth cell-specific alpha-defensin 1;PDBTitle: the solution structure of defa1, a highly potent antimicrobial peptide2 from the horse