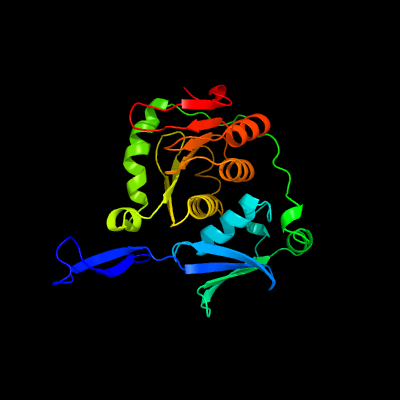
| 1 | c4pdeA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein fdhd;
PDBTitle: crystal structure of fdhd in complex with gdp
|
|
|
|
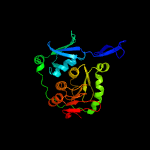
| 2 | d2pw9a1
|
|
 |
100.0 |
25 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:FdhD/NarQ |
|
|
|
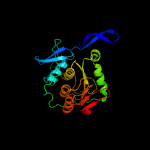
| 3 | d2nu7a1
|
|
 |
89.1 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
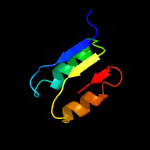
| 4 | d1oi7a1
|
|
 |
88.9 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
| 5 | c6hxqA_
|
|
 |
68.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:citryl-coa synthetase small subunit;
PDBTitle: structure of citryl-coa synthetase from hydrogenobacter thermophilus
|
|
|
|
| 6 | c3mwdB_
|
|
 |
65.9 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
|
|
|
| 7 | c3evnA_
|
|
 |
64.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
|
|
|
|
| 8 | c2duwA_
|
|
 |
62.6 |
19 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:putative coa-binding protein;
PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
|
|
|
|
| 9 | d1iuka_
|
|
 |
60.3 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
| 10 | c1y88A_
|
|
 |
56.4 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af1548;
PDBTitle: crystal structure of protein of unknown function af1548
|
|
|
|
| 11 | c4h3vA_
|
|
 |
53.6 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:oxidoreductase domain protein;
PDBTitle: crystal structure of oxidoreductase domain protein from kribbella2 flavida
|
|
|
|
| 12 | c3ff4A_
|
|
 |
52.8 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
|
|
|
| 13 | c3dtyA_
|
|
 |
52.6 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
|
|
|
|
| 14 | d1y88a2
|
|
 |
51.8 |
22 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:MRR-like |
|
|
|
| 15 | d2csua1
|
|
 |
51.5 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
| 16 | d1y81a1
|
|
 |
50.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
| 17 | c4xb1B_
|
|
 |
50.4 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:319aa long hypothetical homoserine dehydrogenase;
PDBTitle: hyperthermophilic archaeal homoserine dehydrogenase in complex with2 nadph
|
|
|
|
| 18 | c5yeeA_
|
|
 |
45.9 |
24 |
PDB header:structural protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of lokiprofilin1/rabbit actin complex
|
|
|
|
| 19 | d1j5pa4
|
|
 |
44.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 20 | c1oi7A_
|
|
 |
43.2 |
25 |
PDB header:synthetase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
|
|
|
|
| 21 | d2zdra2 |
|
not modelled |
42.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
|
|
| 22 | d1q3qa2 |
|
not modelled |
41.8 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
|
|
| 23 | c1y4hA_ |
|
not modelled |
41.8 |
7 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:cysteine protease;
PDBTitle: wild type staphopain-staphostatin complex
|
|
|
| 24 | c3euwB_ |
|
not modelled |
41.4 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
|
|
| 25 | d1mxsa_ |
|
not modelled |
41.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 26 | d1vlia2 |
|
not modelled |
40.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
|
|
| 27 | c2v4iA_ |
|
not modelled |
40.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate n-acetyltransferase 2 alpha chain;
PDBTitle: structure of a novel n-acyl-enzyme intermediate of an n-2 terminal nucleophile (ntn) hydrolase, oat2
|
|
|
| 28 | c4gmfD_ |
|
not modelled |
39.6 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:yersiniabactin biosynthetic protein ybtu;
PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
|
|
|
| 29 | c6daoB_ |
|
not modelled |
39.2 |
6 |
PDB header:lyase
Chain: B: PDB Molecule:trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;
PDBTitle: nahe wt selenomethionine
|
|
|
| 30 | c2hwgA_ |
|
not modelled |
39.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
|
|
|
| 31 | d1assa_ |
|
not modelled |
38.6 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
|
|
| 32 | c6oviA_ |
|
not modelled |
38.3 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 33 | c1xuzA_ |
|
not modelled |
37.9 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
|
|
| 34 | d2d59a1 |
|
not modelled |
37.7 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 35 | d1gmla_ |
|
not modelled |
37.5 |
16 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
|
|
| 36 | d1kzyc2 |
|
not modelled |
37.4 |
17 |
Fold:BRCT domain
Superfamily:BRCT domain
Family:53BP1 |
|
|
| 37 | c3oa0B_ |
|
not modelled |
37.4 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lipopolysaccaride biosynthesis protein wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from thermus2 thermophilus in complex with nad and udp-glcnaca
|
|
|
| 38 | c3g8rA_ |
|
not modelled |
36.8 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
|
|
| 39 | c3f4lF_ |
|
not modelled |
36.0 |
30 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative oxidoreductase yhhx;
PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
|
|
|
| 40 | c2hmcA_ |
|
not modelled |
35.3 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
|
|
|
| 41 | d1e0ta2 |
|
not modelled |
35.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 42 | d1g8ma2 |
|
not modelled |
34.6 |
25 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
|
|
| 43 | c4qccA_ |
|
not modelled |
34.2 |
18 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 44 | c6iauB_ |
|
not modelled |
33.9 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:amine dehydrogenase;
PDBTitle: amine dehydrogenase from cystobacter fuscus in complex with nadp+ and2 cyclohexylamine
|
|
|
| 45 | c2qneA_ |
|
not modelled |
33.5 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of putative methyltransferase (zp_00558420.1) from2 desulfitobacterium hafniense y51 at 2.30 a resolution
|
|
|
| 46 | d1h6da1 |
|
not modelled |
33.3 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 47 | d1euca1 |
|
not modelled |
32.8 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 48 | c3ip3D_ |
|
not modelled |
32.7 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, putative;
PDBTitle: structure of putative oxidoreductase (tm_0425) from thermotoga2 maritima
|
|
|
| 49 | d1wpga2 |
|
not modelled |
32.1 |
24 |
Fold:HAD-like
Superfamily:HAD-like
Family:Meta-cation ATPase, catalytic domain P |
|
|
| 50 | c3oa2B_ |
|
not modelled |
32.1 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from pseudomonas2 aeruginosa in complex with nad at 1.5 angstrom resolution
|
|
|
| 51 | c6ncsB_ |
|
not modelled |
32.0 |
21 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:n-acetylneuraminic acid (sialic acid) synthetase;
PDBTitle: crystal structure of n-acetylneuraminic acid (sialic acid) synthetase2 from leptospira borgpetersenii serovar hardjo-bovis in complex with3 citrate
|
|
|
| 52 | c2yw3E_ |
|
not modelled |
31.9 |
33 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
|
|
| 53 | c5uibA_ |
|
not modelled |
31.7 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase protein;
PDBTitle: crystal structure of an oxidoreductase from agrobacterium radiobacter2 in complex with nad+, l-tartaric acid and magnesium
|
|
|
| 54 | c3wb9A_ |
|
not modelled |
31.6 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diaminopimelate dehydrogenase;
PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
|
|
|
| 55 | c2o48X_ |
|
not modelled |
31.0 |
20 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
|
|
| 56 | d1cv8a_ |
|
not modelled |
31.0 |
4 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Papain-like |
|
|
| 57 | c5b3uB_ |
|
not modelled |
29.7 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 58 | c5avoA_ |
|
not modelled |
29.3 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: crystal structure of the reduced form of homoserine dehydrogenase from2 sulfolobus tokodaii.
|
|
|
| 59 | c3lciA_ |
|
not modelled |
28.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
|
|
| 60 | c6jnkA_ |
|
not modelled |
28.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arabinose 1-dehydrogenase (nad(p)(+));
PDBTitle: crystal structure of azospirillum brasilense l-arabinose 1-2 dehydrogenase (nadp-bound form)
|
|
|
| 61 | d2a6na1 |
|
not modelled |
28.8 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 62 | c3e82A_ |
|
not modelled |
28.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from2 klebsiella pneumoniae
|
|
|
| 63 | c5kt0A_ |
|
not modelled |
28.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: dihydrodipicolinate reductase from the industrial and evolutionarily2 important cyanobacteria anabaena variabilis.
|
|
|
| 64 | c5uf2A_ |
|
not modelled |
28.6 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose 5 phosphate isomerase a from neisseria2 gonorrhoeae
|
|
|
| 65 | c1j3wB_ |
|
not modelled |
28.6 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:giding protein-mglb;
PDBTitle: structure of gliding protein-mglb from thermus thermophilus hb8
|
|
|
| 66 | c2fpgA_ |
|
not modelled |
28.5 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa synthetase in2 complex with gdp
|
|
|
| 67 | d1a6db2 |
|
not modelled |
28.3 |
22 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
|
|
| 68 | c3wycB_ |
|
not modelled |
28.0 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:meso-diaminopimelate d-dehydrogenase;
PDBTitle: structure of a meso-diaminopimelate dehydrogenase in complex with nadp
|
|
|
| 69 | c3e0fA_ |
|
not modelled |
27.8 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent phosphoesterase;
PDBTitle: crystal structure of a putative metal-dependent phosphoesterase2 (bad_1165) from bifidobacterium adolescentis atcc 15703 at 2.40 a3 resolution
|
|
|
| 70 | c3aq1B_ |
|
not modelled |
27.8 |
20 |
PDB header:chaperone
Chain: B: PDB Molecule:thermosome subunit;
PDBTitle: open state monomer of a group ii chaperonin from methanococcoides2 burtonii
|
|
|
| 71 | c4mxeA_ |
|
not modelled |
27.3 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetyltransferase esco1;
PDBTitle: human esco1 (eco1/ctf7 ortholog), acetyltransferase domain in complex2 with acetyl-coa
|
|
|
| 72 | c2glxD_ |
|
not modelled |
26.9 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
|
|
| 73 | c3u3xJ_ |
|
not modelled |
26.8 |
21 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from sinorhizobium2 meliloti 1021
|
|
|
| 74 | c3p9ee_ |
|
not modelled |
26.7 |
18 |
PDB header:chaperone
Chain: E: PDB Molecule:
PDBTitle: the crystal structure of yeast cct reveals intrinsic asymmetry of2 eukaryotic cytosolic chaperonins
|
|
|
| 75 | c4fb5A_ |
|
not modelled |
26.5 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of a probable oxidoreduxtase protein
|
|
|
| 76 | c2nu8D_ |
|
not modelled |
26.4 |
19 |
PDB header:ligase
Chain: D: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
|
|
|
| 77 | c4hktA_ |
|
not modelled |
26.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol 2-dehydrogenase;
PDBTitle: crystal structure of a putative myo-inositol dehydrogenase from2 sinorhizobium meliloti 1021 (target psi-012312)
|
|
|
| 78 | c1zh8B_ |
|
not modelled |
25.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
|
|
| 79 | c3nt5B_ |
|
not modelled |
25.8 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
|
|
| 80 | c2ia5C_ |
|
not modelled |
25.2 |
29 |
PDB header:transferase
Chain: C: PDB Molecule:polynucleotide kinase;
PDBTitle: t4 polynucleotide kinase/phosphatase with bound sulfate and magnesium.
|
|
|
| 81 | d1vz6a_ |
|
not modelled |
25.2 |
21 |
Fold:DmpA/ArgJ-like
Superfamily:DmpA/ArgJ-like
Family:ArgJ-like |
|
|
| 82 | d1hl2a_ |
|
not modelled |
25.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 83 | c2ixaA_ |
|
not modelled |
24.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
|
|
| 84 | c6o6dA_ |
|
not modelled |
24.6 |
35 |
PDB header:ligase
Chain: A: PDB Molecule:translation initiation factor if-3;
PDBTitle: n-terminal domain of translation initiation factor if-3 from2 helicobacter pylori
|
|
|
| 85 | c4hbrD_ |
|
not modelled |
24.5 |
17 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:putative periplasmic protein;
PDBTitle: crystal structure of a putative periplasmic proteins (bacegg_01429)2 from bacteroides eggerthii dsm 20697 at 2.40 a resolution
|
|
|
| 86 | c6hxjB_ |
|
not modelled |
24.5 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:atp-citrate lyase alpha-subunit;
PDBTitle: structure of atp citrate lyase from chlorobium limicola in complex2 with citrate and coenzyme a.
|
|
|
| 87 | d1f74a_ |
|
not modelled |
24.2 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 88 | c5yabD_ |
|
not modelled |
24.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:scyllo-inositol dehydrogenase with l-glucose dehydrogenase
PDBTitle: crystal structure of scyllo-inositol dehydrogenase with l-glucose2 dehydrogenase activity
|
|
|
| 89 | d1ebfa1 |
|
not modelled |
23.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 90 | c3oqbF_ |
|
not modelled |
23.7 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
|
|
| 91 | c3e18A_ |
|
not modelled |
23.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
|
|
| 92 | d2ioja1 |
|
not modelled |
23.6 |
15 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:DRTGG domain |
|
|
| 93 | d2nvwa1 |
|
not modelled |
23.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 94 | c1xeaD_ |
|
not modelled |
23.3 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
|
|
| 95 | c1lc3A_ |
|
not modelled |
23.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biliverdin reductase a;
PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
|
|
|
| 96 | c1vliA_ |
|
not modelled |
22.9 |
14 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
|
|
| 97 | c3fkkA_ |
|
not modelled |
22.9 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 98 | c4i7vD_ |
|
not modelled |
22.8 |
25 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: agrobacterium tumefaciens dhdps with pyruvate
|
|
|
| 99 | c5z2fA_ |
|
not modelled |
22.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
|
|
|
| 100 | d1lc0a1 |
|
not modelled |
22.8 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 101 | c2ehhE_ |
|
not modelled |
22.7 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
|
|
|
| 102 | c5n1wA_ |
|
not modelled |
22.5 |
22 |
PDB header:cell cycle
Chain: A: PDB Molecule:xeco2;
PDBTitle: structure of xeco2 acetyltransferase domain bound to k105-coa2 conjugate
|
|
|
| 103 | c3e9mC_ |
|
not modelled |
22.4 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 104 | c4hadD_ |
|
not modelled |
22.2 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
|
|
|
| 105 | c6hxiD_ |
|
not modelled |
22.2 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:succinyl-coa ligase (adp-forming) subunit alpha;
PDBTitle: structure of atp citrate lyase from methanothrix soehngenii in complex2 with citrate and coenzyme a
|
|
|
| 106 | c3pueA_ |
|
not modelled |
22.2 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
| 107 | c2v9dB_ |
|
not modelled |
22.0 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
|
|
| 108 | c3bt3B_ |
|
not modelled |
21.7 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:glyoxalase-related enzyme, arac type;
PDBTitle: crystal structure of a glyoxalase-related enzyme from clostridium2 phytofermentans
|
|
|
| 109 | c3cprB_ |
|
not modelled |
21.6 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthetase;
PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
|
|
|
| 110 | c3ec7C_ |
|
not modelled |
21.4 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
|
|
| 111 | c4ah7C_ |
|
not modelled |
21.2 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: structure of wild type stapylococcus aureus n-acetylneuraminic acid2 lyase in complex with pyruvate
|
|
|
| 112 | c1m0sA_ |
|
not modelled |
20.7 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: northeast structural genomics consortium (nesg id ir21)
|
|
|
| 113 | c5ui3C_ |
|
not modelled |
20.4 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from chlamydomonas reinhardtii
|
|
|
| 114 | c3kyeC_ |
|
not modelled |
20.4 |
23 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:roadblock/lc7 domain, robl_lc7;
PDBTitle: crystal structure of roadblock/lc7 domain from streptomyces2 avermitilis
|
|
|