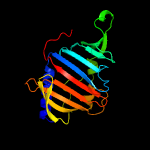
| 1 | d2byoa1
|
|
 |
100.0 |
93 |
Fold:LolA-like prokaryotic lipoproteins and lipoprotein localization factors
Superfamily:Prokaryotic lipoproteins and lipoprotein localization factors
Family:LppX-like |
|
|
|
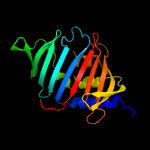
| 2 | c3mhaB_
|
|
 |
100.0 |
32 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:lipoprotein lprg;
PDBTitle: crystal structure of lprg from mycobacterium tuberculosis bound to pim
|
|
|
|
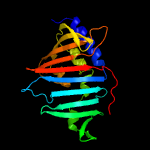
| 3 | c4qa8A_
|
|
 |
100.0 |
31 |
PDB header:lipid transport
Chain: A: PDB Molecule:putative lipoprotein lprf;
PDBTitle: crystal structure of lprf from mycobacterium bovis
|
|
|
|
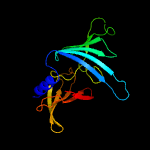
| 4 | c3buuB_
|
|
 |
97.8 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized lola superfamily protein ne2245;
PDBTitle: crystal structure of lola superfamily protein ne2245 from2 nitrosomonas europaea
|
|
|
|
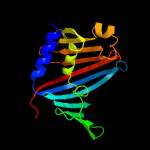
| 5 | c4mxtA_
|
|
 |
97.1 |
14 |
PDB header:protein transport
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an outer-membrane lipoprotein carrier protein2 (bacuni_04723) from bacteroides uniformis atcc 8492 at 1.40 a3 resolution
|
|
|
|
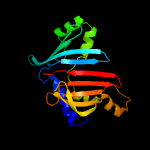
| 6 | c6in8A_
|
|
 |
96.2 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:sigma factor algu regulatory protein mucb;
PDBTitle: crystal structure of mucb
|
|
|
|
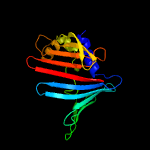
| 7 | c2v43A_
|
|
 |
94.2 |
15 |
PDB header:regulator
Chain: A: PDB Molecule:sigma-e factor regulatory protein rseb;
PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
|
|
|
|
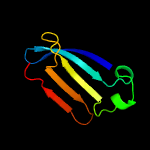
| 8 | c4z48B_
|
|
 |
90.1 |
22 |
PDB header:structural biology, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf1329 family protein (despig_00262) from2 desulfovibrio piger atcc 29098 at 1.75 a resolution
|
|
|
|
| 9 | c3bk5A_
|
|
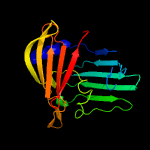 |
82.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative outer membrane lipoprotein-sorting protein;
PDBTitle: crystal structure of putative outer membrane lipoprotein-sorting2 protein domain from vibrio parahaemolyticus
|
|
|
|
| 10 | c3woaA_
|
|
 |
76.5 |
7 |
PDB header:dna binding protein, sugar binding prote
Chain: A: PDB Molecule:repressor protein ci, maltose-binding periplasmic protein;
PDBTitle: crystal structure of lambda repressor (1-45) fused with maltose-2 binding protein
|
|
|
|
| 11 | c4mjsQ_
|
|
 |
49.3 |
25 |
PDB header:transferase/protein binding
Chain: Q: PDB Molecule:protein kinase c zeta type;
PDBTitle: crystal structure of a pb1 complex
|
|
|
|
| 12 | c2w7qB_
|
|
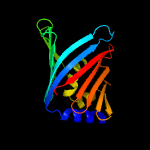 |
45.9 |
10 |
PDB header:protein transport
Chain: B: PDB Molecule:outer-membrane lipoprotein carrier protein;
PDBTitle: structure of pseudomonas aeruginosa lola
|
|
|
|
| 13 | c6mitC_
|
|
 |
21.0 |
30 |
PDB header:lipid transport
Chain: C: PDB Molecule:lipopolysaccharide export system protein lptc;
PDBTitle: lptbfgc from enterobacter cloacae
|
|
|
|
| 14 | c5b3zB_
|
|
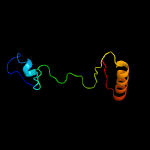 |
16.3 |
15 |
PDB header:isomerase,sugar binding protein
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase nima-interacting 1,
PDBTitle: crystal structure of hpin1 ww domain (5-39) fused with maltose-binding2 protein
|
|
|
|
| 15 | c5az3A_
|
|
 |
16.0 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type transporter, periplasmic component;
PDBTitle: crystal structure of heme binding protein hmut
|
|
|
|
| 16 | c3p8dB_
|
|
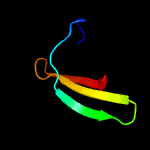 |
15.0 |
9 |
PDB header:protein binding
Chain: B: PDB Molecule:medulloblastoma antigen mu-mb-50.72;
PDBTitle: crystal structure of the second tudor domain of human phf20 (homodimer2 form)
|
|
|
|
| 17 | c2ldmA_
|
|
 |
14.5 |
9 |
PDB header:transcription/protein binding
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of human phf20 tudor2 domain bound to a p53 segment2 containing a dimethyllysine analog p53k370me2
|
|
|
|
| 18 | c4zv4C_
|
|
 |
12.9 |
44 |
PDB header:translation
Chain: C: PDB Molecule:tse6;
PDBTitle: structure of tse6 in complex with ef-tu
|
|
|
|
| 19 | d2hqxa1
|
|
 |
11.1 |
12 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
|
|
|
| 20 | c2hqxB_
|
|
 |
11.1 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:p100 co-activator tudor domain;
PDBTitle: crystal structure of human p100 tudor domain conserved2 region
|
|
|
|
| 21 | d1wmxa_ |
|
not modelled |
8.9 |
32 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:Family 30 carbohydrate binding module, CBM30 (PKD repeat) |
|
|
| 22 | c5o60Y_ |
|
not modelled |
8.8 |
16 |
PDB header:ribosome
Chain: Y: PDB Molecule:50s ribosomal protein l28;
PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
|
|
|
| 23 | c2equA_ |
|
not modelled |
7.9 |
6 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
|
|
| 24 | c6gwjD_ |
|
not modelled |
7.6 |
19 |
PDB header:rna binding protein
Chain: D: PDB Molecule:ekc/keops complex subunit gon7;
PDBTitle: protein complex
|
|
|
| 25 | c2e76D_ |
|
not modelled |
7.5 |
4 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
|
|
| 26 | c3qiiA_ |
|
not modelled |
6.9 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:phd finger protein 20;
PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
|
|
|
| 27 | c2k9yB_ |
|
not modelled |
6.5 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
|
|
| 28 | c2k9yA_ |
|
not modelled |
6.5 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
|
|
|
| 29 | c1nbjA_ |
|
not modelled |
6.5 |
38 |
PDB header:plant protein
Chain: A: PDB Molecule:cycloviolacin o1;
PDBTitle: high-resolution solution structure of cycloviolacin o1
|
|
|
| 30 | d1nbja_ |
|
not modelled |
6.5 |
38 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Cyclotides
Family:Cycloviolacin |
|
|
| 31 | d1r17a1 |
|
not modelled |
6.5 |
15 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Bacterial adhesins
Family:Fibrinogen-binding domain |
|
|
| 32 | c2lojA_ |
|
not modelled |
6.2 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cytoplasmic protein;
PDBTitle: solution nmr structure of tstm1273 from salmonella typhimurium lt2,2 nesg target stt322, csgid target idp01027 and ocsp target tstm1273
|
|
|
| 33 | c1vb8A_ |
|
not modelled |
6.0 |
38 |
PDB header:plant protein
Chain: A: PDB Molecule:viola hederacea root peptide 1;
PDBTitle: solution structure of vhr1, the first cyclotide from root2 tissue
|
|
|
| 34 | d1vb8a_ |
|
not modelled |
6.0 |
38 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Cyclotides
Family:Cycloviolacin |
|
|
| 35 | c4e6nB_ |
|
not modelled |
5.9 |
25 |
PDB header:protein binding
Chain: B: PDB Molecule:methyltransferase type 12;
PDBTitle: crystal structure of bacterial pnkp-c/hen1-n heterodimer
|
|
|
| 36 | c2jraB_ |
|
not modelled |
5.4 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein rpa2121;
PDBTitle: a novel domain-swapped solution nmr structure of protein rpa2121 from2 rhodopseudomonas palustris. northeast structural genomics target rpt6
|
|
|
| 37 | c4q5wB_ |
|
not modelled |
5.4 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:maternal protein tudor;
PDBTitle: crystal structure of extended-tudor 9 of drosophila melanogaster
|
|
|












































































































































































