| 1 | c6eqoB_
|
|
 |
100.0 |
20 |
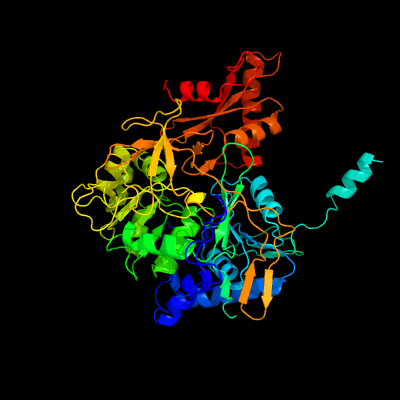
PDB header:oxidoreductase
Chain: B: PDB Molecule:acetyl-coenzyme a synthetase;
PDBTitle: tri-functional propionyl-coa synthase of erythrobacter sp. nap1 with2 bound nadp+ and phosphomethylphosphonic acid adenylate ester
|
|
|
|
| 2 | c5gxdA_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:amp-dependent synthetase and ligase;
PDBTitle: structure of acryloyl-coa lyase prpe from dinoroseobacter shibae dfl2 12
|
|
|
|
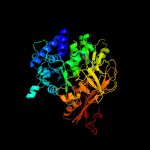
| 3 | c5es8A_
|
|
 |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:linear gramicidin synthetase subunit a;
PDBTitle: crystal structure of the initiation module of lgra in the thiolation2 state
|
|
|
|
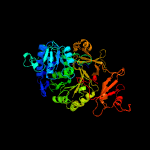
| 4 | c6p1jA_
|
|
 |
100.0 |
24 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:txo2;
PDBTitle: the structure of condensation and adenylation domains of teixobactin-2 producing nonribosomal peptide synthetase txo2 serine module
|
|
|
|
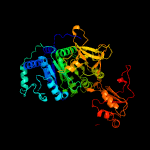
| 5 | c5u89A_
|
|
 |
100.0 |
23 |
PDB header:hydrolase/inhibitor
Chain: A: PDB Molecule:amino acid adenylation domain protein;
PDBTitle: crystal structure of a cross-module fragment from the dimodular nrps2 dhbf
|
|
|
|
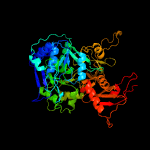
| 6 | d1pg4a_
|
|
 |
100.0 |
17 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
|
| 7 | c5wmmA_
|
|
 |
100.0 |
24 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:nrps;
PDBTitle: crystal structure of an adenylation domain interrupted by a2 methylation domain (ama4) from nonribosomal peptide synthetase tios
|
|
|
|
| 8 | c5ja2A_
|
|
 |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:enterobactin synthase component f;
PDBTitle: entf, a terminal nonribosomal peptide synthetase module bound to the2 non-native mbth-like protein pa2412
|
|
|
|
| 9 | c5ey8D_
|
|
 |
100.0 |
35 |
PDB header:ligase
Chain: D: PDB Molecule:acyl-coa synthase;
PDBTitle: structure of fadd32 from mycobacterium smegmatis complexed to ampc20
|
|
|
|
| 10 | c5ifiA_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coenzyme a synthetase;
PDBTitle: crystal structure of acetyl-coa synthetase in complex with adenosine-2 5'-propylphosphate from cryptococcus neoformans h99
|
|
|
|
| 11 | c5msdA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carboxylic acid reductase;
PDBTitle: structure of the a domain of carboxylic acid reductase (car) from2 nocardia iowensis in complex with amp and benzoic acid
|
|
|
|
| 12 | c6n8eA_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:holo-obif1;
PDBTitle: crystal structure of holo-obif1, a five domain nonribosomal peptide2 synthetase from burkholderia diffusa
|
|
|
|
| 13 | c2vsqA_
|
|
 |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:surfactin synthetase subunit 3;
PDBTitle: structure of surfactin a synthetase c (srfa-c), a nonribosomal peptide2 synthetase termination module
|
|
|
|
| 14 | d1ry2a_
|
|
 |
100.0 |
18 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
|
| 15 | c4wd1A_
|
|
 |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:acetoacetate-coa ligase;
PDBTitle: acetoacetyl-coa synthetase from streptomyces lividans
|
|
|
|
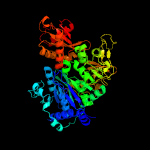
| 16 | c3kxwA_
|
|
 |
100.0 |
33 |
PDB header:ligase
Chain: A: PDB Molecule:saframycin mx1 synthetase b;
PDBTitle: the crystal structure of fatty acid amp ligase from legionella2 pneumophila
|
|
|
|
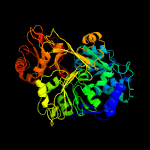
| 17 | c5mstA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioester reductase domain-containing protein;
PDBTitle: structure of the a domain of carboxylic acid reductase (car) from2 segniliparus rugosus in complex with amp and a co-purified carboxylic3 acid
|
|
|
|
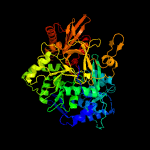
| 18 | c4r0mB_
|
|
 |
100.0 |
25 |
PDB header:ligase
Chain: B: PDB Molecule:mcyg protein;
PDBTitle: structure of mcyg a-pcp complexed with phenylalanyl-adenylate
|
|
|
|
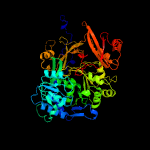
| 19 | c4zxiA_
|
|
 |
100.0 |
20 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:tyrocidine synthetase 3;
PDBTitle: crystal structure of holo-ab3403 a four domain nonribosomal peptide2 synthetase bound to amp and glycine
|
|
|
|
| 20 | c5mssA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioester reductase domain-containing protein;
PDBTitle: structure of the a-pcp didomain of carboxylic acid reductase (car)2 from segniliparus rugosus in complex with amp
|
|
|
|
| 21 | c3gqwB_ |
|
not modelled |
100.0 |
29 |
PDB header:ligase
Chain: B: PDB Molecule:fatty acid amp ligase;
PDBTitle: crystal structure of a fatty acid amp ligase from e. coli with an acyl2 adenylate product bound
|
|
|
| 22 | c4zxjA_ |
|
not modelled |
100.0 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:enterobactin synthase component f;
PDBTitle: crystal structure of holo-entf a nonribosomal peptide synthetase in2 the thioester-forming conformation
|
|
|
| 23 | c3tsyA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase, transferase
Chain: A: PDB Molecule:fusion protein 4-coumarate--coa ligase 1, resveratrol
PDBTitle: 4-coumaroyl-coa ligase::stilbene synthase fusion protein
|
|
|
| 24 | c3e7wA_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--poly(phosphoribitol) ligase subunit 1;
PDBTitle: crystal structure of dlta: implications for the reaction mechanism of2 non-ribosomal peptide synthetase (nrps) adenylation domains
|
|
|
| 25 | c4r0mA_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:mcyg protein;
PDBTitle: structure of mcyg a-pcp complexed with phenylalanyl-adenylate
|
|
|
| 26 | c5ie2A_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:oxalate--coa ligase;
PDBTitle: crystal structure of a plant enzyme
|
|
|
| 27 | c3vnqA_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:nrps adenylation protein cytc1;
PDBTitle: co-crystal structure of nrps adenylation protein cytc1 with atp from2 streptomyces
|
|
|
| 28 | d3cw9a1 |
|
not modelled |
100.0 |
20 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 29 | d1mdba_ |
|
not modelled |
100.0 |
18 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 30 | c4wv3A_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase/ligase inhibitor
Chain: A: PDB Molecule:anthranilate-coa ligase;
PDBTitle: crystal structure of the anthranilate coa ligase auaeii in complex2 with anthranoyl-amp
|
|
|
| 31 | c4eatB_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:benzoate-coenzyme a ligase;
PDBTitle: crystal structure of a benzoate coenzyme a ligase
|
|
|
| 32 | c5x8gA_ |
|
not modelled |
100.0 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:2-succinylbenzoate--coa ligase;
PDBTitle: binary complex structure of a double mutant i454ra456k of o-2 succinylbenzoate coa synthetase (mene) from bacillus subtilis bound3 with its product analogue osb-ncoa at 1.90 angstrom
|
|
|
| 33 | c3etcB_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:amp-binding protein;
PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
|
|
|
| 34 | c3ni2A_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:4-coumarate:coa ligase;
PDBTitle: crystal structures and enzymatic mechanisms of a populus tomentosa 4-2 coumarate:coa ligase
|
|
|
| 35 | c4dg9A_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase/inhibitor
Chain: A: PDB Molecule:pa1221;
PDBTitle: structure of holo-pa1221, an nrps protein containing adenylation and2 pcp domains bound to vinylsulfonamide inhibitor
|
|
|
| 36 | c3eynB_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:acyl-coenzyme a synthetase acsm2a;
PDBTitle: crystal structure of human acyl-coa synthetase medium-chain2 family member 2a (l64p mutation) in a complex with coa
|
|
|
| 37 | c5wm7A_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:salicylate-amp ligase;
PDBTitle: crystal structure of cahj in complex with amp
|
|
|
| 38 | c3rg2H_ |
|
not modelled |
100.0 |
15 |
PDB header:ligase
Chain: H: PDB Molecule:enterobactin synthase component e (ente), 2,3-dihydro-2,3-
PDBTitle: structure of a two-domain nrps fusion protein containing the ente2 adenylation domain and entb aryl-carrier protein from enterobactin3 biosynthesis
|
|
|
| 39 | c6ozvA_ |
|
not modelled |
100.0 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:txo1;
PDBTitle: the structure of condensation and adenylation domains of teixobactin-2 producing nonribosomal peptide synthetase txo1 serine module in3 complex with amp
|
|
|
| 40 | c5u2aA_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:amp-dependent synthetase and ligase;
PDBTitle: crystal structure of brucella canis acyl-coa synthetase
|
|
|
| 41 | c4dg8A_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:pa1221;
PDBTitle: structure of pa1221, an nrps protein containing adenylation and pcp2 domains
|
|
|
| 42 | c3iteB_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:sidn siderophore synthetase;
PDBTitle: the third adenylation domain of the fungal sidn non-ribosomal peptide2 synthetase
|
|
|
| 43 | c5aplA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:adenylation domain;
PDBTitle: structure of the adenylation domain thr1 involved in the biosynthesis2 of 4-chlorothreonine in streptomyces sp. oh-5093, apo structure
|
|
|
| 44 | c6ac3B_ |
|
not modelled |
100.0 |
18 |
PDB header:luminescent protein
Chain: B: PDB Molecule:red-bioluminescence eliciting luciferase;
PDBTitle: structure of a natural red emitting luciferase from phrixothrix hirtus2 (p3121 crystal form)
|
|
|
| 45 | c4oxiA_ |
|
not modelled |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:enterobactin synthetase component f-related protein;
PDBTitle: crystal structure of vibrio cholerae adenylation domain alme in2 complex with glycyl-adenosine-5'-phosphate
|
|
|
| 46 | c3r44A_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:fatty acyl coa synthetase fadd13 (fatty-acyl-coa
PDBTitle: mycobacterium tuberculosis fatty acyl coa synthetase
|
|
|
| 47 | c6ijbA_ |
|
not modelled |
100.0 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:amp-binding domain protein;
PDBTitle: structure of 3-methylmercaptopropionate coa ligase mutant k523a in2 complex with amp and mmpa
|
|
|
| 48 | c2d1tA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:luciferin 4-monooxygenase;
PDBTitle: crystal structure of the thermostable japanese firefly2 luciferase red-color emission s286n mutant complexed with3 high-energy intermediate analogue
|
|
|
| 49 | c6h1bA_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:fatty acid coa ligase;
PDBTitle: structure of amide bond synthetase mcba k483a mutant from2 marinactinospora thermotolerans
|
|
|
| 50 | c4d56A_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:apnaa1;
PDBTitle: understanding bi-specificity of a-domains
|
|
|
| 51 | c4ir7A_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:long chain fatty acid coa ligase fadd10;
PDBTitle: crystal structure of mtb fadd10 in complex with dodecanoyl-amp
|
|
|
| 52 | c6akdA_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:amp-dependent synthetase and ligase;
PDBTitle: crystal structure of idnl7
|
|
|
| 53 | d1amua_ |
|
not modelled |
100.0 |
17 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 54 | c5keiA_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:2,3-dihydroxybenzoate-amp ligase;
PDBTitle: mycobacterium smegmatis mbta apo structure
|
|
|
| 55 | c1amuB_ |
|
not modelled |
100.0 |
18 |
PDB header:peptide synthetase
Chain: B: PDB Molecule:gramicidin synthetase 1;
PDBTitle: phenylalanine activating domain of gramicidin synthetase 12 in a complex with amp and phenylalanine
|
|
|
| 56 | c5e7qB_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:acyl-coa synthetase;
PDBTitle: acyl-coa synthetase ptma2 from streptomyces platensis
|
|
|
| 57 | c2v7bB_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:benzoate-coenzyme a ligase;
PDBTitle: crystal structures of a benzoate coa ligase from2 burkholderia xenovorans lb400
|
|
|
| 58 | d1v25a_ |
|
not modelled |
100.0 |
20 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 59 | c4fuqD_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: D: PDB Molecule:malonyl coa synthetase;
PDBTitle: crystal structure of apo matb from rhodopseudomonas palustris
|
|
|
| 60 | c5jjqB_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:amp-dependent synthetase and ligase;
PDBTitle: crystal structure of idnl1
|
|
|
| 61 | c3g7sA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:long-chain-fatty-acid--coa ligase (fadd-1);
PDBTitle: crystal structure of a long-chain-fatty-acid-coa ligase2 (fadd1) from archaeoglobus fulgidus
|
|
|
| 62 | c3dhvA_ |
|
not modelled |
100.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-poly(phosphoribitol) ligase;
PDBTitle: crystal structure of dlta protein in complex with d-alanine2 adenylate
|
|
|
| 63 | c4gr5B_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:non-ribosomal peptide synthetase;
PDBTitle: crystal structure of slgn1deltaasub in complex with ampcpp
|
|
|
| 64 | c3l8cA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--poly(phosphoribitol) ligase subunit 1;
PDBTitle: structure of probable d-alanine--poly(phosphoribitol) ligase subunit-12 from streptococcus pyogenes
|
|
|
| 65 | d1lcia_ |
|
not modelled |
100.0 |
17 |
Fold:Acetyl-CoA synthetase-like
Superfamily:Acetyl-CoA synthetase-like
Family:Acetyl-CoA synthetase-like |
|
|
| 66 | c5buqA_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:2-succinylbenzoate--coa ligase;
PDBTitle: unliganded form of o-succinylbenzoate coenzyme a synthetase (mene)2 from bacillus subtilis, solved at 1.98 angstroms
|
|
|
| 67 | c3nyrA_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:malonyl-coa ligase;
PDBTitle: malonyl-coa ligase ternary product complex with malonyl-coa and amp2 bound
|
|
|
| 68 | c3e53A_ |
|
not modelled |
100.0 |
54 |
PDB header:ligase
Chain: A: PDB Molecule:fatty-acid-coa ligase fadd28;
PDBTitle: crystal structure of n-terminal domain of a fatty acyl amp2 ligase faal28 from mycobacterium tuberculosis
|
|
|
| 69 | c3iplB_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:2-succinylbenzoate--coa ligase;
PDBTitle: crystal structure of o-succinylbenzoic acid-coa ligase from2 staphylococcus aureus subsp. aureus mu50
|
|
|
| 70 | c4w8oA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:luciferase-like enzymeamp-coa-ligase;
PDBTitle: structure of the luciferase-like enzyme from the nonluminescent2 zophobas morio mealworm
|
|
|
| 71 | c5c5hA_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:2-succinylbenzoate--coa ligase;
PDBTitle: r195k e. coli mene with bound osb-ams
|
|
|
| 72 | c5jjpB_ |
|
not modelled |
100.0 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:nonribosomal peptide synthase;
PDBTitle: crystal structure of cmis6
|
|
|
| 73 | c3o82B_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:peptide arylation enzyme;
PDBTitle: structure of base n-terminal domain from acinetobacter baumannii bound2 to 5'-o-[n-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
|
|
|
| 74 | c3qyaA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:luciferase;
PDBTitle: crystal structure of a red-emitter mutant of lampyris turkestanicus2 luciferase
|
|
|
| 75 | c5n81B_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:tyrocidine synthase 1;
PDBTitle: crystal structure of an engineered tyca variant in complex with an o-2 propargyl-beta-tyr-amp analog
|
|
|
| 76 | c4lgcA_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:bile acid-coenzyme a ligase;
PDBTitle: crystal structure of a bile acid-coenzyme a ligase (baib) from2 clostridium scindens (vpi 12708) at 2.19 a resolution
|
|
|
| 77 | c3o82A_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:peptide arylation enzyme;
PDBTitle: structure of base n-terminal domain from acinetobacter baumannii bound2 to 5'-o-[n-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine
|
|
|
| 78 | c3wv4B_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:non-ribosomal peptide synthetase;
PDBTitle: crystal structure of vinn
|
|
|
| 79 | c3ivrA_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:putative long-chain-fatty-acid coa ligase;
PDBTitle: crystal structure of putative long-chain-fatty-acid coa ligase from2 rhodopseudomonas palustris cga009
|
|
|
| 80 | c3t5cA_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:probable chain-fatty-acid-coa ligase fadd13;
PDBTitle: crystal structure of n-terminal domain of facl13 from mycobacterium2 tuberculosis in different space group c2
|
|
|
| 81 | c5oe3C_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:anthranilate--coa ligase;
PDBTitle: crystal structure of the n-terminal domain of pqsa in complex with2 anthraniloyl-amp (crystal form 1)
|
|
|
| 82 | c5burB_ |
|
not modelled |
100.0 |
26 |
PDB header:ligase
Chain: B: PDB Molecule:2-succinylbenzoate--coa ligase;
PDBTitle: o-succinylbenzoate coenzyme a synthetase (mene) from bacillus2 subtilis, in complex with atp and magnesium ion
|
|
|
| 83 | c6abhG_ |
|
not modelled |
100.0 |
17 |
PDB header:luminescent protein
Chain: G: PDB Molecule:red-bioluminescence eliciting luciferase;
PDBTitle: structure of a natural red emitting luciferase from phrixothrix hirtus2 (p1 crystal form)
|
|
|
| 84 | c5jjpC_ |
|
not modelled |
100.0 |
24 |
PDB header:ligase
Chain: C: PDB Molecule:nonribosomal peptide synthase;
PDBTitle: crystal structure of cmis6
|
|
|
| 85 | c2y4oA_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:phenylacetate-coenzyme a ligase;
PDBTitle: crystal structure of paak2 in complex with phenylacetyl adenylate
|
|
|
| 86 | c2y27B_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:phenylacetate-coenzyme a ligase;
PDBTitle: crystal structure of paak1 in complex with atp from burkholderia2 cenocepacia
|
|
|
| 87 | c4gs5A_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:acyl-coa synthetase (amp-forming)/amp-acid ligase ii-like
PDBTitle: the crystal structure of acyl-coa synthetase (amp-forming)/amp-acid2 ligase ii-like protein from dyadobacter fermentans dsm 18053
|
|
|
| 88 | c3qovD_ |
|
not modelled |
100.0 |
15 |
PDB header:ligase
Chain: D: PDB Molecule:phenylacetate-coenzyme a ligase;
PDBTitle: crystal structure of a hypothetical acyl-coa ligase (bt_0428) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
|
|
|
| 89 | c3hguB_ |
|
not modelled |
100.0 |
12 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:ehpf;
PDBTitle: structure of phenazine antibiotic biosynthesis protein
|
|
|
| 90 | c3laxA_ |
|
not modelled |
99.6 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:phenylacetate-coenzyme a ligase;
PDBTitle: the crystal structure of a domain of phenylacetate-coenzyme2 a ligase from bacteroides vulgatus atcc 8482
|
|
|
| 91 | c5kodA_ |
|
not modelled |
99.4 |
9 |
PDB header:ligase
Chain: A: PDB Molecule:indole-3-acetic acid-amido synthetase gh3.5;
PDBTitle: crystal structure of gh3.5 acyl acid amido synthetase from arabidopsis2 thaliana
|
|
|
| 92 | c4eplA_ |
|
not modelled |
99.3 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:jasmonic acid-amido synthetase jar1;
PDBTitle: crystal structure of arabidopsis thaliana gh3.11 (jar1) in complex2 with ja-ile
|
|
|
| 93 | c4b2gB_ |
|
not modelled |
99.0 |
11 |
PDB header:signaling protein
Chain: B: PDB Molecule:gh3-1 auxin conjugating enzyme;
PDBTitle: crystal structure of an indole-3-acetic acid amido synthase from vitis2 vinifera involved in auxin homeostasis
|
|
|
| 94 | c6avhA_ |
|
not modelled |
98.2 |
14 |
PDB header:ligase, plant protein
Chain: A: PDB Molecule:gh3.15 acyl acid amido synthetase;
PDBTitle: gh3.15 acyl acid amido synthetase
|
|
|
| 95 | c4ewvB_ |
|
not modelled |
98.2 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:4-substituted benzoates-glutamate ligase gh3.12;
PDBTitle: crystal structure of gh3.12 in complex with ampcpp
|
|
|
| 96 | c6hnuA_ |
|
not modelled |
65.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:aromatic amino acid aminotransferase i;
PDBTitle: crystal structure of the aminotransferase aro8 from c. albicans with2 ligands
|
|
|
| 97 | c4j5uB_ |
|
not modelled |
60.1 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: x-ray crystal structure of a serine hydroxymethyltransferase with2 covalently bound plp from rickettsia rickettsii str. sheila smith
|
|
|
| 98 | c4je5C_ |
|
not modelled |
58.5 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:aromatic/aminoadipate aminotransferase 1;
PDBTitle: crystal structure of the aromatic aminotransferase aro8, a putative2 alpha-aminoadipate aminotransferase in saccharomyces cerevisiae
|
|
|
| 99 | c2dkjB_ |
|
not modelled |
40.2 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of t.th.hb8 serine hydroxymethyltransferase
|
|
|
| 100 | d1kl1a_ |
|
not modelled |
38.2 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 101 | c4wxfC_ |
|
not modelled |
34.7 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of l-serine hydroxymethyltransferase in complex with2 glycine
|
|
|
| 102 | d1nula_ |
|
not modelled |
33.8 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 103 | c6raoI_ |
|
not modelled |
32.3 |
12 |
PDB header:virus like particle
Chain: I: PDB Molecule:afp11;
PDBTitle: cryo-em structure of the anti-feeding prophage (afp) baseplate, 6-fold2 symmetrised
|
|
|
| 104 | c4z1oB_ |
|
not modelled |
31.2 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosyltransferase;
PDBTitle: hypoxanthine-guanine-xanthine phosphoribosyltransferase (hgxprt) from2 sulfolobus solfataricus in complex with alpha-3 phosphoribosylpyrophosphoric acid (prpp) and magnesium
|
|
|
| 105 | d2ayia1 |
|
not modelled |
31.0 |
17 |
Fold:Thermophilic metalloprotease-like
Superfamily:Thermophilic metalloprotease-like
Family:Thermophilic metalloprotease (M29) |
|
|
| 106 | c3h7fB_ |
|
not modelled |
30.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase 1;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 mycobacterium tuberculosis
|
|
|
| 107 | d1ejia_ |
|
not modelled |
30.2 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
|
|
| 108 | d1uj4a2 |
|
not modelled |
28.8 |
19 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
|
|
| 109 | c2ypdB_ |
|
not modelled |
27.7 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable jmjc domain-containing histone demethylation prot
PDBTitle: crystal structure of the jumonji domain of human jumonji domain2 containing 1c protein
|
|
|
| 110 | c5vogA_ |
|
not modelled |
27.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphoribosyltransferase;
PDBTitle: crystal structure of a hypothetical protein from neisseria gonorrhoeae2 with bound ppgpp
|
|
|
| 111 | c3d6kB_ |
|
not modelled |
26.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:putative aminotransferase;
PDBTitle: the crystal structure of a putative aminotransferase from2 corynebacterium diphtheriae
|
|
|
| 112 | c4trbA_ |
|
not modelled |
25.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:purine phosphoribosyltransferase (gpt-1);
PDBTitle: sulfolobus solfataricus adenine phosphoribosyltransferase
|
|
|
| 113 | c4bheI_ |
|
not modelled |
22.0 |
15 |
PDB header:transferase
Chain: I: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: methanococcus jannaschii serine hydroxymethyl-transferase2 in complex with plp
|
|
|
| 114 | c5f1yA_ |
|
not modelled |
21.9 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:mccc family protein;
PDBTitle: crystal structure of ba3275, the member of s66 family of serine2 peptidases
|
|
|