| 1 | c3w7bB_
|
|
 |
100.0 |
58 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase from thermus2 thermophilus hb8
|
|
|
|
| 2 | c3o1lB_
|
|
 |
100.0 |
45 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
|
|
|
| 3 | c3obiC_
|
|
 |
100.0 |
45 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
|
|
|
| 4 | c3louB_
|
|
 |
100.0 |
43 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
|
|
|
| 5 | c3n0vD_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
|
|
|
| 6 | c3nrbD_
|
|
 |
100.0 |
46 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
|
|
|
| 7 | c3aufA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:glycinamide ribonucleotide transformylase 1;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 symbiobacterium toebii
|
|
|
|
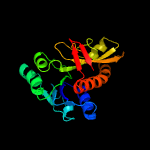
| 8 | c2ywrA_
|
|
 |
100.0 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of gar transformylase from aquifex aeolicus
|
|
|
|
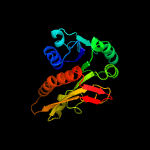
| 9 | d1jkxa_
|
|
 |
100.0 |
27 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
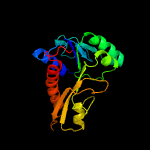
| 10 | c3p9xB_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
|
|
|
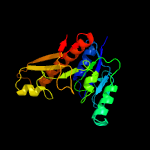
| 11 | c3tqrA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
|
|
|
| 12 | c3av3A_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 geobacillus kaustophilus
|
|
|
|
| 13 | d1meoa_
|
|
 |
100.0 |
29 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 14 | c3dcjA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide formyltransferase
PDBTitle: crystal structure of glycinamide formyltransferase (purn) from2 mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-3 tetrahydrofolic acid derivative
|
|
|
|
| 15 | c3kcqA_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
|
|
|
| 16 | c4ds3A_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 brucella melitensis
|
|
|
|
| 17 | c4s1nA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
|
|
|
|
| 18 | c5cjjA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
|
| 19 | d1fmta2
|
|
 |
100.0 |
22 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 20 | d1s3ia2
|
|
 |
100.0 |
21 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
|
| 21 | c1z7eC_ |
|
not modelled |
100.0 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
|
|
| 22 | c1fmtA_ |
|
not modelled |
100.0 |
22 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
|
|
| 23 | d2blna2 |
|
not modelled |
100.0 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 24 | c3tqqA_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: structure of the methionyl-trna formyltransferase (fmt) from coxiella2 burnetii
|
|
|
| 25 | d2bw0a2 |
|
not modelled |
100.0 |
20 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 26 | c3rfoA_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
|
|
| 27 | c1yrwA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
|
|
| 28 | c5uaiA_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methionyl-trna formyltransferase from pseudomonas2 aeruginosa
|
|
|
| 29 | c1s3iA_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:10-formyltetrahydrofolate dehydrogenase;
PDBTitle: crystal structure of the n terminal hydrolase domain of 10-2 formyltetrahydrofolate dehydrogenase
|
|
|
| 30 | c3q0iA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from vibrio cholerae
|
|
|
| 31 | c4nv1D_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:formyltransferase;
PDBTitle: crystal structure of a 4-n formyltransferase from francisella2 tularensis
|
|
|
| 32 | c4pzuF_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: F: PDB Molecule:uncharacterized protein rv3404c/mt3512;
PDBTitle: crystal structure of a putative uncharacterize protein rv3404c and2 likely sugar n-formyltransferase from mycobacterium tuberculosis
|
|
|
| 33 | c6nbpA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:n-formyltransferase;
PDBTitle: crystal structure of a sugar n-formyltransferase from the plant2 pathogen pantoea ananatis
|
|
|
| 34 | c5vytD_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-
PDBTitle: crystal structure of the wbkc n-formyltransferase (f142a variant) from2 brucella melitensis
|
|
|
| 35 | c4yfvA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:viof;
PDBTitle: x-ray structure of the 4-n-formyltransferase viof from providencia2 alcalifaciens o30
|
|
|
| 36 | c6ci2A_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:formyltransferase psej;
PDBTitle: crystal structure of the formyltransferase psej from anoxybacillus2 kamchatkensis
|
|
|
| 37 | c5uimB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:formyltransferase;
PDBTitle: x-ray structure of the fdtf n-formyltransferase from salmonella2 enteric o60 in complex with folinic acid and tdp-qui3n
|
|
|
| 38 | c4lxuB_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:wlard, a sugar 3n-formyl transferase;
PDBTitle: dtdp-fuc3n and 5-n-formyl-thf
|
|
|
| 39 | d1zgha2 |
|
not modelled |
100.0 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 40 | c1zghA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from clostridium thermocellum
|
|
|
| 41 | c4xd0A_ |
|
not modelled |
99.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:tdp-3-aminoquinovose-n-formyltransferase;
PDBTitle: x-ray structure of the n-formyltransferase qdtf from providencia2 alcalifaciens
|
|
|
| 42 | d1zpva1 |
|
not modelled |
99.6 |
21 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
|
|
| 43 | c1u8sB_ |
|
not modelled |
99.4 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
|
|
| 44 | d1u8sa1 |
|
not modelled |
99.4 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
|
|
| 45 | d1u8sa2 |
|
not modelled |
99.2 |
14 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
|
|
| 46 | c2nyiB_ |
|
not modelled |
99.0 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria sulphuraria
|
|
|
| 47 | c6culG_ |
|
not modelled |
98.2 |
26 |
PDB header:transferase
Chain: G: PDB Molecule:pyoverdine synthetase f;
PDBTitle: pvdf of pyoverdin biosynthesis is a structurally unique n10-2 formyltetrahydrofolate-dependent formyltransferase
|
|
|
| 48 | c3p96A_ |
|
not modelled |
97.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase serb;
PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
|
|
|
| 49 | c3ibwA_ |
|
not modelled |
97.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp pyrophosphokinase of2 chlorobium tepidum. northeast structural genomics consortium target3 ctr148a
|
|
|
| 50 | d1ygya3 |
|
not modelled |
97.0 |
22 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
|
|
| 51 | d1sc6a3 |
|
not modelled |
97.0 |
18 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
|
|
| 52 | c1ygyA_ |
|
not modelled |
96.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
|
|
|
| 53 | c5uscB_ |
|
not modelled |
96.1 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase tyra from bacillus2 anthracis in complex with nad and l-tyrosine
|
|
|
| 54 | c1y7pB_ |
|
not modelled |
95.8 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein af1403;
PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
|
|
|
| 55 | d1ru8a_ |
|
not modelled |
95.6 |
19 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 56 | c2lvwA_ |
|
not modelled |
95.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme 1 small subunit;
PDBTitle: solution nmr studies of the dimeric regulatory subunit ilvn of the2 e.coli acetohydroxyacid synthase i (ahas i)
|
|
|
| 57 | c2fgcA_ |
|
not modelled |
94.9 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
|
|
| 58 | d2fgca2 |
|
not modelled |
94.8 |
24 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
|
|
| 59 | c2pc6C_ |
|
not modelled |
94.8 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
|
|
| 60 | c3a14B_ |
|
not modelled |
94.5 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
|
|
|
| 61 | c2f1fA_ |
|
not modelled |
94.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of acetohydroxyacid2 synthase isozyme iii from e. coli
|
|
|
| 62 | d2pc6a2 |
|
not modelled |
93.9 |
22 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
|
|
| 63 | c1ni5A_ |
|
not modelled |
93.7 |
15 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative cell cycle protein mesj;
PDBTitle: structure of the mesj pp-atpase from escherichia coli
|
|
|
| 64 | d2f1fa1 |
|
not modelled |
93.5 |
21 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
|
|
| 65 | c6dzsD_ |
|
not modelled |
93.5 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:homoserine dehydrogenase;
PDBTitle: mycobacterial homoserine dehydrogenase thra in complex with nadp
|
|
|
| 66 | d1f0ka_ |
|
not modelled |
93.1 |
16 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
|
|
| 67 | d1r0ka2 |
|
not modelled |
93.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 68 | d1xi8a3 |
|
not modelled |
92.8 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 69 | c2e21A_ |
|
not modelled |
92.5 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
|
|
|
| 70 | d2d13a1 |
|
not modelled |
91.9 |
27 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 71 | c2f06B_ |
|
not modelled |
91.6 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
|
|
| 72 | c1ofgF_ |
|
not modelled |
91.6 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
|
|
| 73 | c1h6dL_ |
|
not modelled |
91.5 |
13 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
|
|
| 74 | d2f06a2 |
|
not modelled |
91.0 |
23 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
|
|
| 75 | c3luyA_ |
|
not modelled |
90.7 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:probable chorismate mutase;
PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
|
|
|
| 76 | c5g2rA_ |
|
not modelled |
90.3 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin biosynthesis protein cnx1;
PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana
|
|
|
| 77 | d1ni5a1 |
|
not modelled |
90.1 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
|
|
| 78 | d1wu2a3 |
|
not modelled |
90.0 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 79 | c2jcyA_ |
|
not modelled |
89.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium tuberculosis
|
|
|
| 80 | d2ftsa3 |
|
not modelled |
89.4 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 81 | c3a2kB_ |
|
not modelled |
88.3 |
15 |
PDB header:ligase/rna
Chain: B: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils complexed with trna
|
|
|
| 82 | c2qmxB_ |
|
not modelled |
88.2 |
12 |
PDB header:ligase
Chain: B: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
|
|
|
| 83 | c3mtjA_ |
|
not modelled |
88.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
|
|
|
| 84 | c3s2uA_ |
|
not modelled |
88.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine--n-acetylmuramyl-(pentapeptide)
PDBTitle: crystal structure of the pseudomonas aeruginosa murg:udp-glcnac2 substrate complex
|
|
|
| 85 | d2f06a1 |
|
not modelled |
87.8 |
22 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
|
|
| 86 | d1wy5a1 |
|
not modelled |
87.3 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
|
|
| 87 | c2nqqA_ |
|
not modelled |
87.1 |
21 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
|
|
| 88 | c1r0lD_ |
|
not modelled |
86.7 |
25 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
|
|
|
| 89 | c5udwB_ |
|
not modelled |
86.5 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:lactate racemization operon protein lare;
PDBTitle: lare, a sulfur transferase involved in synthesis of the cofactor for2 lactate racemase, in complex with nickel
|
|
|
| 90 | c2fu3A_ |
|
not modelled |
85.9 |
21 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
|
|
| 91 | c2hmaA_ |
|
not modelled |
85.7 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:probable trna (5-methylaminomethyl-2-thiouridylate)-
PDBTitle: the crystal structure of trna (5-methylaminomethyl-2-thiouridylate)-2 methyltransferase trmu from streptococcus pneumoniae
|
|
|
| 92 | c4gmfD_ |
|
not modelled |
85.2 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:yersiniabactin biosynthetic protein ybtu;
PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
|
|
|
| 93 | d1phza1 |
|
not modelled |
85.0 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
|
|
| 94 | c3e9mC_ |
|
not modelled |
84.8 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
|
|
| 95 | c5b3uB_ |
|
not modelled |
84.6 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 96 | c1uz5A_ |
|
not modelled |
84.2 |
16 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
|
|
| 97 | c2derA_ |
|
not modelled |
84.1 |
15 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna-specific 2-thiouridylase mnma;
PDBTitle: cocrystal structure of an rna sulfuration enzyme mnma and2 trna-glu in the initial trna binding state
|
|
|
| 98 | c3l76B_ |
|
not modelled |
83.7 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
|
|
| 99 | c3bl5E_ |
|
not modelled |
83.4 |
16 |
PDB header:hydrolase
Chain: E: PDB Molecule:queuosine biosynthesis protein quec;
PDBTitle: crystal structure of quec from bacillus subtilis: an enzyme2 involved in preq1 biosynthesis
|
|
|
| 100 | c4oh7B_ |
|
not modelled |
81.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from brucella2 melitensis
|
|
|
| 101 | d1uz5a3 |
|
not modelled |
81.2 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 102 | d2hmfa3 |
|
not modelled |
79.4 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
|
|
| 103 | c2nvwB_ |
|
not modelled |
79.3 |
8 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
|
|
| 104 | c2q8nB_ |
|
not modelled |
78.3 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of glucose-6-phosphate isomerase (ec 5.3.1.9)2 (tm1385) from thermotoga maritima at 1.82 a resolution
|
|
|
| 105 | c5l78A_ |
|
not modelled |
76.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-aminoadipic semialdehyde synthase, mitochondrial;
PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
|
|
|
| 106 | d1q0qa2 |
|
not modelled |
76.5 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 107 | c3s1tB_ |
|
not modelled |
76.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: structure of the regulatory domain of aspartokinase (rv3709c; ak-beta)2 in complex with threonine from mycobacterium tuberculosis
|
|
|
| 108 | d2nvwa1 |
|
not modelled |
76.0 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 109 | d1jsca3 |
|
not modelled |
75.9 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
|
|
| 110 | c3u3xJ_ |
|
not modelled |
75.9 |
22 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from sinorhizobium2 meliloti 1021
|
|
|
| 111 | c4gqaC_ |
|
not modelled |
75.6 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad binding oxidoreductase;
PDBTitle: crystal structure of nad binding oxidoreductase from klebsiella2 pneumoniae
|
|
|
| 112 | c3q2kB_ |
|
not modelled |
74.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
|
|
| 113 | c3rbvA_ |
|
not modelled |
74.8 |
21 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
|
|
| 114 | c5fa0B_ |
|
not modelled |
74.3 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:putative n-acetyl glucosaminyl transferase;
PDBTitle: the structure of the beta-3-deoxy-d-manno-oct-2-ulosonic acid2 transferase domain from wbbb
|
|
|
| 115 | c1ybaC_ |
|
not modelled |
74.2 |
30 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: the active form of phosphoglycerate dehydrogenase
|
|
|
| 116 | c4ycsC_ |
|
not modelled |
73.2 |
13 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative lipoprotein from peptoclostridium2 difficile 630 (fragment)
|
|
|
| 117 | c3gd5D_ |
|
not modelled |
73.2 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase from gloeobacter2 violaceus
|
|
|
| 118 | d1j8yf2 |
|
not modelled |
73.0 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 119 | c1xeaD_ |
|
not modelled |
72.7 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
|
|
| 120 | d1mvla_ |
|
not modelled |
70.9 |
19 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
|
|