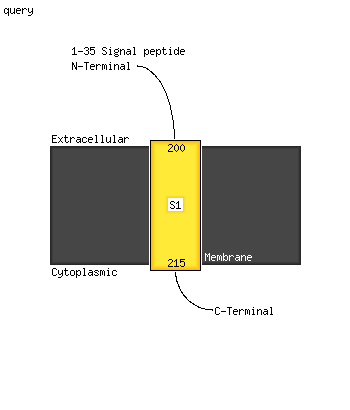
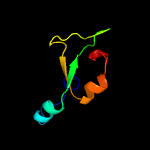
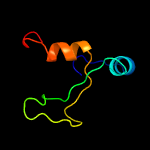
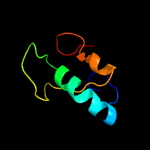
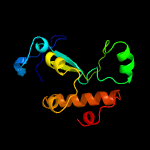
1 c5vgbA_
99.0
25
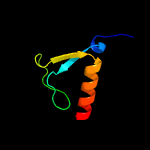
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: crispr-associated endonuclease cas9;PDBTitle: crystal structure of nmecas9 hnh domain bound to anti-crispr acriic1
2 c5axwA_
98.5
19
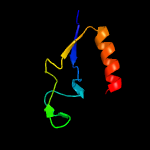
PDB header: hydrolase/rna/dnaChain: A: PDB Molecule: crispr-associated endonuclease cas9;PDBTitle: crystal structure of staphylococcus aureus cas9 in complex with sgrna2 and target dna (ttgggt pam)
3 c4ogeA_
97.8
40
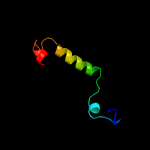
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease domain protein;PDBTitle: crystal structure of the type ii-c cas9 enzyme from actinomyces2 naeslundii
4 c2qgpA_
97.5
22
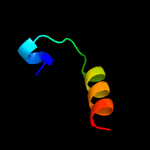
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium target3 gmr87.
5 c4cmqB_
96.8
30
PDB header: hydrolaseChain: B: PDB Molecule: crispr-associated endonuclease cas9/csn1;PDBTitle: crystal structure of mn-bound s.pyogenes cas9
6 d1ouoa_
94.1
19
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Endonuclease I
7 c5mkwA_
92.2
19
PDB header: hydrolaseChain: A: PDB Molecule: dna annealing helicase and endonuclease zranb3;PDBTitle: crystal structure of the human zranb3 hnh domain
8 c5h0mA_
85.2
28
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: crystal structure of deep-sea thermophilic bacteriophage gve2 hnh2 endonuclease with zinc ion
9 c5fhzF_
37.8
18
PDB header: oxidoreductaseChain: F: PDB Molecule: aldehyde dehydrogenase family 1 member a3;PDBTitle: human aldehyde dehydrogenase 1a3 complexed with nad(+) and retinoic2 acid
10 d1jmxa1
28.6
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
11 c2o2qA_
27.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: formyltetrahydrofolate dehydrogenase;PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp
12 c4qyjD_
27.5
20
PDB header: oxidoreductaseChain: D: PDB Molecule: aldehyde dehydrogenase;PDBTitle: structure of phenylacetaldehyde dehydrogenase from pseudomonas putida2 s12
13 c5b2oA_
26.4
50
PDB header: hydrolase/rna/dnaChain: A: PDB Molecule: crispr-associated endonuclease cas9;PDBTitle: crystal structure of francisella novicida cas9 in complex with sgrna2 and target dna (tgg pam)
14 c2m76A_
24.0
32
PDB header: signaling proteinChain: A: PDB Molecule: carnitine o-palmitoyltransferase 1, brain isoform;PDBTitle: structure of the regulatory domain of human brain carnitine2 palmitoyltransferase 1
15 d1pbya1
23.9
4
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
16 c2hqhE_
23.4
64
PDB header: structural protein, protein bindingChain: E: PDB Molecule: restin;PDBTitle: crystal structure of p150glued and clip-170
17 c3e2uF_
23.4
64
PDB header: protein bindingChain: F: PDB Molecule: cap-gly domain-containing linker protein 1;PDBTitle: crystal structure of the zink-knuckle 2 domain of human clip-170 in2 complex with cap-gly domain of human dynactin-1 (p150-glued)
18 c3e2uE_
23.4
64
PDB header: protein bindingChain: E: PDB Molecule: cap-gly domain-containing linker protein 1;PDBTitle: crystal structure of the zink-knuckle 2 domain of human clip-170 in2 complex with cap-gly domain of human dynactin-1 (p150-glued)
19 d1bxsa_
23.1
22
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
20 c3vz0B_
22.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nad-dependent aldehyde dehydrogenase;PDBTitle: structural insights into cofactor and substrate selection by gox0499
21 c2hg2A_
not modelled
21.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase a;PDBTitle: structure of lactaldehyde dehydrogenase
22 c2mnjA_
not modelled
21.7
26
PDB header: protein bindingChain: A: PDB Molecule: tpr repeat-containing protein associated with hsp90;PDBTitle: nmr solution structure of the yeast pih1 and tah1 c-terminal domains2 complex
23 c4o5hD_
not modelled
21.2
22
PDB header: oxidoreductaseChain: D: PDB Molecule: phenylacetaldehyde dehydrogenase;PDBTitle: x-ray crystal structure of a putative phenylacetaldehyde dehydrogenase2 from burkholderia cenocepacia
24 c3b4wA_
not modelled
21.1
27
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of mycobacterium tuberculosis aldehyde dehydrogenase2 complexed with nad+
25 c3e2uH_
not modelled
20.7
64
PDB header: protein bindingChain: H: PDB Molecule: cap-gly domain-containing linker protein 1;PDBTitle: crystal structure of the zink-knuckle 2 domain of human clip-170 in2 complex with cap-gly domain of human dynactin-1 (p150-glued)
26 d1o9ja_
not modelled
20.4
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
27 c4pxlB_
not modelled
20.3
33
PDB header: oxidoreductaseChain: B: PDB Molecule: cytosolic aldehyde dehydrogenase rf2c;PDBTitle: structure of zm aldh2-3 (rf2c) in complex with nad
28 c4lihG_
not modelled
19.9
16
PDB header: oxidoreductaseChain: G: PDB Molecule: gamma-glutamyl-gamma-aminobutyraldehyde dehydrogenase;PDBTitle: the crystal structure of gamma-glutamyl-gamma-aminobutyraldehyde2 dehydrogenase from burkholderia cenocepacia j2315
29 c5mz5A_
not modelled
19.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: aldh21);PDBTitle: crystal structure of aldehyde dehydrogenase 21 (aldh21) from2 physcomitrella patens in its apoform
30 c5iuuA_
not modelled
18.2
20
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase family protein;PDBTitle: crystal structure of indole-3-acetaldehyde dehydrogenase in apo form
31 c3efvC_
not modelled
16.8
16
PDB header: oxidoreductaseChain: C: PDB Molecule: putative succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of a putative succinate-semialdehyde dehydrogenase2 from salmonella typhimurium lt2 with bound nad
32 c3rh9A_
not modelled
16.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase (nad(p)(+));PDBTitle: the crystal structure of oxidoreductase from marinobacter aquaeolei
33 c3r64A_
not modelled
15.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nad dependent benzaldehyde dehydrogenase;PDBTitle: crystal structure of a nad-dependent benzaldehyde dehydrogenase from2 corynebacterium glutamicum
34 c4jz6A_
not modelled
15.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: salicylaldehyde dehydrogenase nahf;PDBTitle: crystal structure of a salicylaldehyde dehydrogenase from pseudomonas2 putida g7 complexed with salicylaldehyde
35 c5vbfH_
not modelled
14.7
16
PDB header: oxidoreductaseChain: H: PDB Molecule: nad-dependent succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of succinate semialdehyde dehydrogenase from2 burkholderia vietnamiensis
36 c2ve5H_
not modelled
14.1
11
PDB header: oxidoreductaseChain: H: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystallographic structure of betaine aldehyde2 dehydrogenase from pseudomonas aeruginosa
37 d1pgl22
not modelled
14.0
22
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP
38 d2f2ha3
not modelled
13.9
9
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Putative glucosidase YicI, domain 3
39 c4h7nA_
not modelled
13.9
9
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: the structure of putative aldehyde dehydrogenase puta from anabaena2 variabilis.
40 c5f7sA_
not modelled
13.8
26
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 31;PDBTitle: cycloalternan-degrading enzyme from trueperella pyogenes
41 c1xsiF_
not modelled
13.6
11
PDB header: hydrolaseChain: F: PDB Molecule: putative family 31 glucosidase yici;PDBTitle: structure of a family 31 alpha glycosidase
42 d1ny722
not modelled
13.5
17
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP
43 c3ifgH_
not modelled
13.1
16
PDB header: oxidoreductaseChain: H: PDB Molecule: succinate-semialdehyde dehydrogenase (nadp+);PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2
44 d1wnda_
not modelled
13.0
16
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
45 d2fnoa1
not modelled
12.7
23
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
46 c3pqaA_
not modelled
12.7
9
PDB header: oxidoreductaseChain: A: PDB Molecule: lactaldehyde dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase gapn2 from methanocaldococcus jannaschii dsm 2661
47 c4pt3C_
not modelled
12.4
24
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: nadph complex structure of aldehyde dehydrogenase from bacillus cereus
48 c3rkoE_
not modelled
12.1
31
PDB header: oxidoreductaseChain: E: PDB Molecule: nadh-quinone oxidoreductase subunit a;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
49 c3rkoA_
not modelled
12.1
31
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase subunit a;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
50 c4go4E_
not modelled
12.1
16
PDB header: oxidoreductaseChain: E: PDB Molecule: putative gamma-hydroxymuconic semialdehyde dehydrogenase;PDBTitle: crystal structure of pnpe in complex with nicotinamide adenine2 dinucleotide
51 c5u0mB_
not modelled
12.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: n-succinylglutamate 5-semialdehyde dehydrogenase;PDBTitle: fatty aldehyde dehydrogenase from marinobacter aquaeolei vt8 and2 cofactor complex
52 c5tjrE_
not modelled
11.7
13
PDB header: oxidoreductaseChain: E: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: x-ray crystal structure of a methylmalonate semialdehyde dehydrogenase2 from pseudomonas sp. aac
53 c4dngB_
not modelled
11.4
13
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized aldehyde dehydrogenase aldy;PDBTitle: crystal structure of putative aldehyde dehydrogenase from bacillus2 subtilis subsp. subtilis str. 168
54 c3prlD_
not modelled
11.3
9
PDB header: oxidoreductaseChain: D: PDB Molecule: nadp-dependent glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-phosphate2 dehydrogenase from bacillus halodurans c-125
55 c4pxnB_
not modelled
11.2
11
PDB header: oxidoreductaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: structure of zm aldh7 in complex with nad
56 c3vdoB_
not modelled
11.2
18
PDB header: dna binding protein/protein bindingChain: B: PDB Molecule: anti-sigma-k factor rska;PDBTitle: structure of extra-cytoplasmic function(ecf) sigma factor sigk in2 complex with its negative regulator rska from mycobacterium3 tuberculosis
57 c3rosA_
not modelled
10.9
27
PDB header: oxidoreductaseChain: A: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 lactobacillus acidophilus
58 c3ek1C_
not modelled
10.8
13
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from brucella2 melitensis biovar abortus 2308
59 c5j6bB_
not modelled
10.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from burkholderia2 thailandensis in covelent complex with nadph
60 c4e4gF_
not modelled
10.3
13
PDB header: oxidoreductaseChain: F: PDB Molecule: methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of putative methylmalonate-semialdehyde2 dehydrogenase from sinorhizobium meliloti 1021
61 c4itaA_
not modelled
10.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase;PDBTitle: structure of bacterial enzyme in complex with cofactor
62 c2w8qA_
not modelled
10.2
13
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate-semialdehyde dehydrogenase,PDBTitle: the crystal structure of human ssadh in complex with ssa.
63 d1w7ca3
not modelled
10.2
26
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
64 c5j4aA_
not modelled
9.6
25
PDB header: toxinChain: A: PDB Molecule: trna nuclease cdia;PDBTitle: cdia-ct toxin from burkholderia pseudomallei e479 in complex with2 cognate cdii immunity protein
65 c4yweE_
not modelled
9.3
22
PDB header: oxidoreductaseChain: E: PDB Molecule: putative aldehyde dehydrogenase;PDBTitle: crystal structure of a putative aldehyde dehydrogenase from2 burkholderia cenocepacia
66 c1t90B_
not modelled
9.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: probable methylmalonate-semialdehyde dehydrogenase;PDBTitle: crystal structure of methylmalonate semialdehyde dehydrogenase from2 bacillus subtilis
67 d1o04a_
not modelled
9.3
27
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
68 c2jg7G_
not modelled
9.2
18
PDB header: oxidoreductaseChain: G: PDB Molecule: antiquitin;PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity
69 d1ag8a_
not modelled
9.1
24
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
70 c4xsjA_
not modelled
9.0
53
PDB header: transport proteinChain: A: PDB Molecule: lysozyme,calcium uniporter protein, mitochondrial;PDBTitle: crystal structure of the n-terminal domain of the human mitochondrial2 calcium uniporter fused with t4 lysozyme
71 d1euha_
not modelled
8.9
9
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
72 c3ed6B_
not modelled
8.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
73 c4h73E_
not modelled
8.7
9
PDB header: oxidoreductaseChain: E: PDB Molecule: aldehyde dehydrogenase;PDBTitle: thermostable aldehyde dehydrogenase from pyrobaculum sp. complexed2 with nadp+
74 c5x5uB_
not modelled
8.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: alpha-ketoglutaric semialdehyde dehydrogenase;PDBTitle: crystal strcuture of alpha-ketoglutarate-semialdehyde dehydrogenase2 (kgsadh) complexed with nad
75 c3k2wD_
not modelled
8.5
11
PDB header: oxidoreductaseChain: D: PDB Molecule: betaine-aldehyde dehydrogenase;PDBTitle: crystal structure of betaine-aldehyde dehydrogenase from2 pseudoalteromonas atlantica t6c
76 c4knaA_
not modelled
8.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: n-succinylglutamate 5-semialdehyde dehydrogenase;PDBTitle: crystal structure of an n-succinylglutamate 5-semialdehyde2 dehydrogenase from burkholderia thailandensis
77 c6g72A_
not modelled
8.3
24
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-ubiquinone oxidoreductase chain 3;PDBTitle: mouse mitochondrial complex i in the deactive state
78 d1ky8a_
not modelled
8.2
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
79 c5v89A_
not modelled
8.2
15
PDB header: ligase / protein bindingChain: A: PDB Molecule: dcn1-like protein 4;PDBTitle: structure of dcn4 pony domain bound to cul1 whb
80 c6ghcA_
not modelled
8.2
29
PDB header: hydrolaseChain: A: PDB Molecule: 5-methylcytosine-specific restriction enzyme a;PDBTitle: modification dependent ecokmcra restriction endonuclease
81 c5omtA_
not modelled
7.8
7
PDB header: hydrolaseChain: A: PDB Molecule: nucb;PDBTitle: endonuclease nucb
82 c3topA_
not modelled
7.7
29
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: maltase-glucoamylase, intestinal;PDBTitle: crystral structure of the c-terminal subunit of human maltase-2 glucoamylase in complex with acarbose
83 c5dkyA_
not modelled
7.6
10
PDB header: hydrolaseChain: A: PDB Molecule: alpha glucosidase-like protein;PDBTitle: crystal structure of glucosidase ii alpha subunit (dnj-bound from)
84 c4xpsA_
not modelled
7.6
26
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of the mutant d365a of pedobacter saltans gh312 alpha-galactosidase complexed with p-nitrophenyl-alpha-3 galactopyranoside
85 c5izdE_
not modelled
7.3
13
PDB header: oxidoreductaseChain: E: PDB Molecule: d-glyceraldehyde dehydrogenase (nadp(+));PDBTitle: wild-type glyceraldehyde dehydrogenase from thermoplasma acidophilum2 in complex with nadp
86 c4gaoA_
not modelled
7.3
19
PDB header: ligase/peptideChain: A: PDB Molecule: dcn1-like protein 2;PDBTitle: dcnl complex with n-terminally acetylated nedd8 e2 peptide
87 c3i44A_
not modelled
7.2
22
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from bartonella2 henselae at 2.0a resolution
88 d1a4sa_
not modelled
7.1
18
Fold: ALDH-likeSuperfamily: ALDH-likeFamily: ALDH-like
89 c2bpbB_
not modelled
7.1
16
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
90 c5m73D_
not modelled
7.0
43
PDB header: rna binding proteinChain: D: PDB Molecule: signal recognition particle subunit srp72;PDBTitle: structure of the human srp s domain with srp72 rna-binding domain
91 c5kzxA_
not modelled
7.0
28
PDB header: hydrolaseChain: A: PDB Molecule: lysosomal alpha-glucosidase;PDBTitle: crystal structure of human gaa
92 c4innA_
not modelled
7.0
11
PDB header: transport proteinChain: A: PDB Molecule: hp1028;PDBTitle: protein hp1028 from the human pathogen helicobacter pylori belongs to2 the lipocalin family
93 c6feyH_
not modelled
6.8
33
PDB header: peptide binding proteinChain: H: PDB Molecule: probable insulin-like peptide 5;PDBTitle: crystal structure of drosophila neural ectodermal development factor2 imp-l2 with drosophila dilp5 insulin
94 c6feyF_
not modelled
6.6
33
PDB header: peptide binding proteinChain: F: PDB Molecule: probable insulin-like peptide 5;PDBTitle: crystal structure of drosophila neural ectodermal development factor2 imp-l2 with drosophila dilp5 insulin
95 c2wfuB_
not modelled
6.5
33
PDB header: signaling proteinChain: B: PDB Molecule: probable insulin-like peptide 5 b chain;PDBTitle: crystal structure of dilp5 variant db
96 c2wfvB_
not modelled
6.5
33
PDB header: signaling proteinChain: B: PDB Molecule: probable insulin-like peptide 5 b chain;PDBTitle: crystal structure of dilp5 variant c4
97 c3lppA_
not modelled
6.4
18
PDB header: hydrolaseChain: A: PDB Molecule: sucrase-isomaltase;PDBTitle: crystal complex of n-terminal sucrase-isomaltase with kotalanol
98 d1miwa2
not modelled
6.3
22
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly A polymerase head domain-like
99 c3rlcA_
not modelled
6.2
20
PDB header: structural proteinChain: A: PDB Molecule: a1 protein;PDBTitle: crystal structure of the read-through domain from bacteriophage qbeta2 a1 protein, hexagonal crystal form