1 c2zhxG_
100.0
100
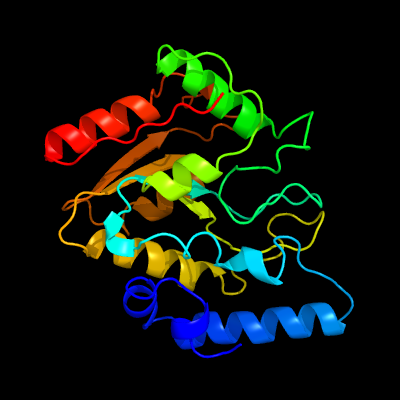
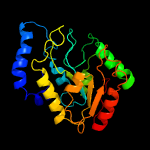
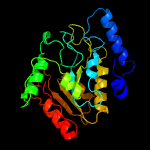
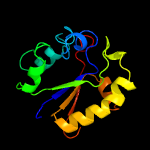
PDB header: hydrolase/hydrolase inhibitorChain: G: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of uracil-dna glycosylase from mycobacterium2 tuberculosis in complex with a proteinaceous inhibitor
2 c3zoqA_
100.0
46
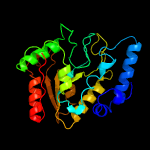
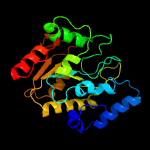
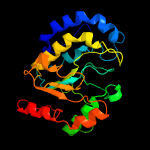
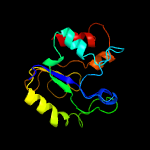
PDB header: hydrolase/viral proteinChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of bsudg-p56 complex
3 c3tr7A_
100.0
41
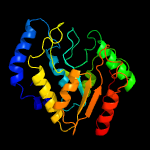
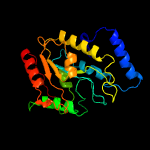
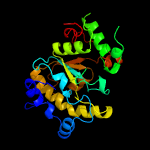
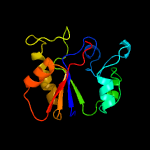
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
4 d1okba_
100.0
39
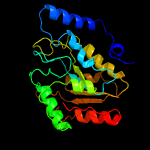
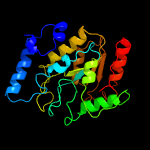
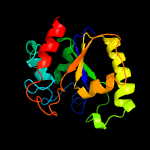
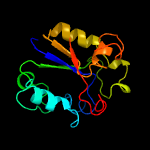
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
5 c3cxmA_
100.0
40
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: leishmania naiffi uracil-dna glycosylase in complex with 5-bromouracil
6 c2booA_
100.0
46
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the crystal structure of uracil-dna n-glycosylase (ung) from2 deinococcus radiodurans.
7 d3euga_
100.0
38
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
8 d2hxma1
100.0
39
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
9 c5x55A_
100.0
29
PDB header: hydrolaseChain: A: PDB Molecule: probable uracil-dna glycosylase;PDBTitle: crystal structure of mimivirus uracil-dna glycosylase
10 d1laue_
100.0
36
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
11 c5nn7A_
100.0
33
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: kshv uracil-dna glycosylase, apo form
12 d2j8xa1
100.0
36
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
13 c2owrD_
100.0
21
PDB header: hydrolaseChain: D: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
14 c5x3hA_
100.0
25
PDB header: dna binding proteinChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the y81g mutant of the ung crystal structure from nitratifractor2 salsuginis
15 c2rbaB_
96.9
11
PDB header: hydrolase/dnaChain: B: PDB Molecule: g/t mismatch-specific thymine dna glycosylase;PDBTitle: structure of human thymine dna glycosylase bound to abasic and2 undamaged dna
16 d1muga_
94.6
17
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Mug-like
17 d1ui0a_
94.4
19
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Mug-like
18 c2d3yA_
94.4
21
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of uracil-dna glycosylase from thermus thermophilus2 hb8
19 c2c2pA_
94.1
14
PDB header: hydrolaseChain: A: PDB Molecule: g/u mismatch-specific dna glycosylase;PDBTitle: the crystal structure of mismatch specific uracil-dna2 glycosylase (mug) from deinococcus radiodurans
20 d1oe4a_
93.4
21
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Single-strand selective monofunctional uracil-DNA glycosylase SMUG1
21 c3ikbB_
not modelled
93.4
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: the structure of a conserved protein from streptococcus2 mutans ua159.
22 c5h93C_
not modelled
92.9
20
PDB header: hydrolaseChain: C: PDB Molecule: geobacter metallireducens smug1;PDBTitle: crystal structure of geobacter metallireducens smug1
23 c4zbzA_
not modelled
90.0
19
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: family 4 uracil-dna glycosylase from sulfolobus tokodaii (free form,2 x-ray wavelength=1.5418)
24 d1vk2a_
not modelled
88.8
20
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Mug-like
25 c6ajrA_
not modelled
88.8
17
PDB header: dna binding proteinChain: A: PDB Molecule: uracil dna glycosylase superfamily protein;PDBTitle: complex form of uracil dna glycosylase x and uracil
26 c2h2wA_
not modelled
55.6
18
PDB header: transferaseChain: A: PDB Molecule: homoserine o-succinyltransferase;PDBTitle: crystal structure of homoserine o-succinyltransferase (ec 2.3.1.46)2 (homoserine o-transsuccinylase) (hts) (tm0881) from thermotoga3 maritima at 2.52 a resolution
27 c5grkA_
not modelled
48.8
100
PDB header: hydrolaseChain: A: PDB Molecule: blr0248 protein;PDBTitle: crystal structure of uracil dna glycosylase -xanthine complex from2 bradyrhizobium diazoefficiens
28 c2l3fA_
not modelled
45.8
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a putative uracil dna glycosylase from2 methanosarcina acetivorans, northeast structural genomics consortium3 target mvr76
29 d2ghra1
not modelled
44.3
21
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: HTS-like
30 c5tbfB_
not modelled
38.3
29
PDB header: translationChain: B: PDB Molecule: translation elongation factor;PDBTitle: crystal structure of semet derivatives of domain2 and domain 3 of rctb
31 c5cjjA_
not modelled
36.3
20
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from campylobacter jejuni subsp. jejuni nctc 11168
32 c3av3A_
not modelled
31.5
26
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 geobacillus kaustophilus
33 d1meoa_
not modelled
30.8
20
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
34 c3p9xB_
not modelled
29.4
24
PDB header: transferaseChain: B: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
35 c2i5kB_
not modelled
29.1
15
PDB header: transferaseChain: B: PDB Molecule: utp--glucose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of ugp1p
36 c2ywrA_
not modelled
28.3
24
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of gar transformylase from aquifex aeolicus
37 c2q4jB_
not modelled
27.7
16
PDB header: transferaseChain: B: PDB Molecule: probable utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
38 c3aufA_
not modelled
27.2
31
PDB header: transferaseChain: A: PDB Molecule: glycinamide ribonucleotide transformylase 1;PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 symbiobacterium toebii
39 c5d88A_
not modelled
26.7
18
PDB header: hydrolaseChain: A: PDB Molecule: predicted protease of the collagenase family;PDBTitle: the structure of the u32 peptidase mk0906
40 c3tqrA_
not modelled
25.3
17
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
41 c4bmaB_
not modelled
25.3
15
PDB header: transferaseChain: B: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: structural of aspergillus fumigatus udp-n-acetylglucosamine2 pyrophosphorylase
42 d2icya2
not modelled
24.0
14
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase
43 c4s1nA_
not modelled
23.7
17
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
44 c3oc9A_
not modelled
23.5
10
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: crystal structure of putative udp-n-acetylglucosamine2 pyrophosphorylase from entamoeba histolytica
45 c4zyaA_
not modelled
22.6
23
PDB header: ligaseChain: A: PDB Molecule: asparagine--trna ligase, cytoplasmic;PDBTitle: the n-terminal extension domain of human asparaginyl-trna synthetase
46 d1r2aa_
not modelled
22.0
28
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
47 d1jkxa_
not modelled
21.3
23
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
48 c2yqsA_
not modelled
21.2
12
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: crystal structure of uridine-diphospho-n-acetylglucosamine2 pyrophosphorylase from candida albicans, in the product-binding form
49 c4ds3A_
not modelled
20.1
23
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 brucella melitensis
50 c4d6uI_
not modelled
19.5
47
PDB header: oxidoreductaseChain: I: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: cytochrome bc1 bound to the 4(1h)-pyridone gsk932121
51 c5zyoD_
not modelled
19.0
30
PDB header: transferaseChain: D: PDB Molecule: ribosomal rna large subunit methyltransferase h;PDBTitle: crystal structure of domain-swapped circular-permuted ybea (cp74) from2 escherichia coli
52 d2hwna1
not modelled
18.7
27
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
53 c3dcjA_
not modelled
17.5
25
PDB header: transferaseChain: A: PDB Molecule: probable 5'-phosphoribosylglycinamide formyltransferasePDBTitle: crystal structure of glycinamide formyltransferase (purn) from2 mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-3 tetrahydrofolic acid derivative
54 c4xzcB_
not modelled
17.4
21
PDB header: rna binding proteinChain: B: PDB Molecule: nucleoprotein;PDBTitle: the crystal structure of kupe virus nucleoprotein
55 c5ubfB_
not modelled
17.2
29
PDB header: dna binding proteinChain: B: PDB Molecule: rctb replication initiator protein;PDBTitle: crystal structure of the rctb domains 2-3, rctb-155-483
56 c4f4mA_
not modelled
17.0
45
PDB header: hydrolase regulatorChain: A: PDB Molecule: papain peptidoglycan amidase effector tse1;PDBTitle: structure of the type vi peptidoglycan amidase effector tse1 (c30a)2 from pseudomonas aeruginosa
57 d1maba2
not modelled
16.7
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase
58 c5jp6A_
not modelled
16.6
30
PDB header: hydrolaseChain: A: PDB Molecule: putative polysaccharide deacetylase;PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
59 d1vm8a_
not modelled
16.4
12
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase
60 c4f0wA_
not modelled
16.2
45
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of type effector tse1 c30a mutant from pseudomonas2 aeruginousa
61 d1jv1a_
not modelled
15.6
12
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: UDP-glucose pyrophosphorylase
62 c1fmtA_
not modelled
15.2
23
PDB header: formyltransferaseChain: A: PDB Molecule: methionyl-trna fmet formyltransferase;PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
63 c4bwsE_
not modelled
15.1
55
PDB header: transcriptionChain: E: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of the heterotrimer of pqbp1, u5-15kd and2 u5-52kd.
64 c4bwqD_
not modelled
14.3
75
PDB header: transcriptionChain: D: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of u5-15kd in a complex with pqbp1
65 c4bwqH_
not modelled
14.3
75
PDB header: transcriptionChain: H: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of u5-15kd in a complex with pqbp1
66 c2y8uA_
not modelled
14.1
19
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: a. nidulans chitin deacetylase
67 c4bwqB_
not modelled
13.9
75
PDB header: transcriptionChain: B: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of u5-15kd in a complex with pqbp1
68 c4bwqF_
not modelled
13.5
75
PDB header: transcriptionChain: F: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of u5-15kd in a complex with pqbp1
69 d1fmta2
not modelled
13.0
22
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
70 d1skyb2
not modelled
12.9
25
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase
71 c3r0qA_
not modelled
12.9
23
PDB header: transferaseChain: A: PDB Molecule: probable protein arginine n-methyltransferase 4.2;PDBTitle: a uniquely open conformation revealed in the crystal structure of2 arabidopsis thaliana protein arginine methyltransferase 10
72 d1fx0a2
not modelled
12.6
25
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase
73 c4zhjA_
not modelled
12.6
15
PDB header: metal binding proteinChain: A: PDB Molecule: mg-chelatase subunit chlh;PDBTitle: crystal structure of the catalytic subunit of magnesium chelatase
74 d2blna2
not modelled
12.3
17
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
75 c4bwsB_
not modelled
12.2
75
PDB header: transcriptionChain: B: PDB Molecule: polyglutamine-binding protein 1;PDBTitle: crystal structure of the heterotrimer of pqbp1, u5-15kd and2 u5-52kd.
76 c1yrwA_
not modelled
12.2
17
PDB header: transferaseChain: A: PDB Molecule: protein arna;PDBTitle: crystal structure of e.coli arna transformylase domain
77 c2p2gD_
not modelled
12.1
16
PDB header: transferaseChain: D: PDB Molecule: ornithine carbamoyltransferase;PDBTitle: crystal structure of ornithine carbamoyltransferase from mycobacterium2 tuberculosis (rv1656): orthorhombic form
78 c5vytD_
not modelled
11.9
22
PDB header: transferaseChain: D: PDB Molecule: gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-PDBTitle: crystal structure of the wbkc n-formyltransferase (f142a variant) from2 brucella melitensis
79 c2q62A_
not modelled
11.7
4
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
80 c3h0dB_
not modelled
11.6
13
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
81 c5h77B_
not modelled
11.5
29
PDB header: signaling protein, immune systemChain: B: PDB Molecule: camp-dependent protein kinase type ii-alpha regulatoryPDBTitle: crystal structure of the pka-protein a fusion protein
82 d2jdia2
not modelled
11.2
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase
83 c3nrbD_
not modelled
11.1
17
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
84 c3u3iA_
not modelled
11.0
22
PDB header: rna binding proteinChain: A: PDB Molecule: nucleocapsid protein;PDBTitle: a rna binding protein from crimean-congo hemorrhagic fever virus
85 c3o1lB_
not modelled
11.0
17
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
86 c4rr2D_
not modelled
10.4
35
PDB header: transferaseChain: D: PDB Molecule: dna primase large subunit;PDBTitle: crystal structure of human primase
87 c4zdtB_
not modelled
10.1
36
PDB header: hydrolaseChain: B: PDB Molecule: structure-specific endonuclease subunit slx4;PDBTitle: crystal structure of the ring finger domain of slx1 in complex with2 the c-terminal domain of slx4
88 d2bw0a2
not modelled
9.9
22
Fold: FormyltransferaseSuperfamily: FormyltransferaseFamily: Formyltransferase
89 d1q3qa2
not modelled
9.6
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
90 c3k1yE_
not modelled
9.5
13
PDB header: oxidoreductaseChain: E: PDB Molecule: oxidoreductase;PDBTitle: x-ray structure of oxidoreductase from corynebacterium diphtheriae.2 orthorombic crystal form, northeast structural genomics consortium3 target cdr100d
91 c3n0vD_
not modelled
9.5
17
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
92 c5oeoC_
not modelled
9.5
30
PDB header: membrane proteinChain: C: PDB Molecule: transient receptor potential cation channel subfamily vPDBTitle: solution structure of the complex of trpv5(655-725) with a calmodulin2 e32q/e68q double mutant
93 c4c9vA_
not modelled
9.5
26
PDB header: signaling proteinChain: A: PDB Molecule: rnf43;PDBTitle: xenopus rnf43 ectodomain in complex with xenopus rspo2 fu1-fu2
94 c5zwlG_
not modelled
9.2
17
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase gamma chain;PDBTitle: crystal structure of the gamma - epsilon complex of photosynthetic2 cyanobacterial f1-atpase
95 d1tska_
not modelled
9.1
39
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
96 c3rfoA_
not modelled
8.9
23
PDB header: transferaseChain: A: PDB Molecule: methionyl-trna formyltransferase;PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
97 c5uaiA_
not modelled
8.9
31
PDB header: transferaseChain: A: PDB Molecule: methionyl-trna formyltransferase;PDBTitle: crystal structure of methionyl-trna formyltransferase from pseudomonas2 aeruginosa
98 c4c84B_
not modelled
8.6
25
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase znrf3;PDBTitle: zebrafish znrf3 ectodomain crystal form i
99 c3q0iA_
not modelled
8.6
29
PDB header: transferaseChain: A: PDB Molecule: methionyl-trna formyltransferase;PDBTitle: methionyl-trna formyltransferase from vibrio cholerae