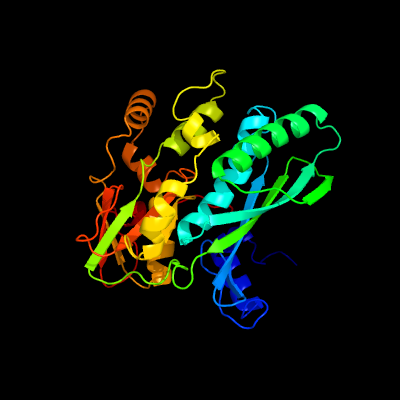
| 1 | c3c9uB_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: aathil complexed with adp and tpp
|
|
|
|
| 2 | c1vqvB_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: crystal structure of thiamine monophosphate kinase (thil)2 from aquifex aeolicus
|
|
|
|
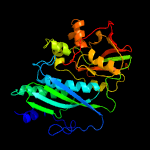
| 3 | c5cm7A_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: structure of thiamine-monophosphate kinase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp) and thiamine3 diphosphate (tpp)
|
|
|
|
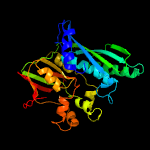
| 4 | c2yxzA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin-monophosphate kinase;
PDBTitle: crystal structure of tt0281 from thermus thermophilus hb8
|
|
|
|
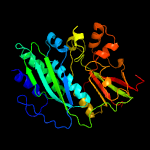
| 5 | c5vk4B_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a phosphoribosylformylglycinamidine cyclo-ligase2 from neisseria gonorrhoeae bound to amppnp and magnesium
|
|
|
|
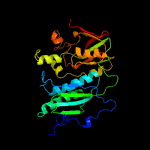
| 6 | c3mcqA_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: crystal structure of thiamine-monophosphate kinase (mfla_0573) from2 methylobacillus flagellatus kt at 1.91 a resolution
|
|
|
|
| 7 | c2btuB_
|
|
 |
100.0 |
16 |
PDB header:synthase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole synthetase;
PDBTitle: crystal structure of phosphoribosylformylglycinamidine cyclo-ligase2 from bacillus anthracis at 2.3a resolution.
|
|
|
|
| 8 | c1cliD_
|
|
 |
100.0 |
18 |
PDB header:ligase
Chain: D: PDB Molecule:protein (phosphoribosyl-aminoimidazole synthetase);
PDBTitle: x-ray crystal structure of aminoimidazole ribonucleotide synthetase2 (purm), from the e. coli purine biosynthetic pathway, at 2.5 a3 resolution
|
|
|
|
| 9 | c3vtiD_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:hydrogenase maturation factor;
PDBTitle: crystal structure of hype-hypf complex
|
|
|
|
| 10 | c2rb9D_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hype protein;
PDBTitle: crystal structure of e.coli hype
|
|
|
|
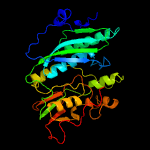
| 11 | c2z1tA_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hydrogenase maturation protein hype
|
|
|
|
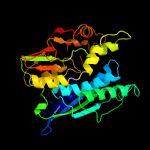
| 12 | c3fd5B_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase 1;
PDBTitle: crystal structure of human selenophosphate synthetase 12 complex with ampcp
|
|
|
|
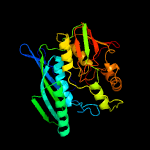
| 13 | c3vysC_
|
|
 |
100.0 |
23 |
PDB header:metal binding protein/transferase
Chain: C: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of the hypc-hypd-hype complex (form i)
|
|
|
|
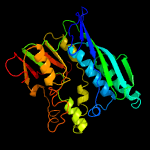
| 14 | c3u0oA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:selenide, water dikinase;
PDBTitle: the crystal structure of selenophosphate synthetase from e. coli
|
|
|
|
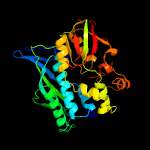
| 15 | c2zodB_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of selenophosphate synthetase from aquifex aeolicus
|
|
|
|
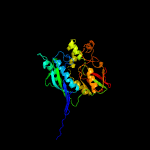
| 16 | c2zauB_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of an n-terminally truncated2 selenophosphate synthetase from aquifex aeolicus
|
|
|
|
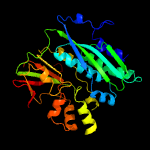
| 17 | c2z01A_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 geobacillus kaustophilus
|
|
|
|
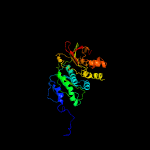
| 18 | c3m84A_
|
|
 |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 francisella tularensis
|
|
|
|
| 19 | c2z1eA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hype from thermococcus kodakaraensis (outward2 form)
|
|
|
|
| 20 | c2v9yA_
|
|
 |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: human aminoimidazole ribonucleotide synthetase
|
|
|
|
| 21 | c5avmE_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: E: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structures of 5-aminoimidazole ribonucleotide (air)2 synthetase, purm, from thermus thermophilus
|
|
|
| 22 | c3ac6A_ |
|
not modelled |
100.0 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase 2;
PDBTitle: crystal structure of purl from thermus thermophilus
|
|
|
| 23 | c5l16A_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:putative selenophosphate synthetase;
PDBTitle: crystal structure of n-terminus truncated selenophosphate synthetase2 from leishmania major
|
|
|
| 24 | c2hs0A_ |
|
not modelled |
100.0 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: t. maritima purl complexed with atp
|
|
|
| 25 | c3d54I_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: I: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: structure of purlqs from thermotoga maritima
|
|
|
| 26 | d3c9ua1 |
|
not modelled |
100.0 |
26 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 27 | c1t3tA_ |
|
not modelled |
100.0 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase;
PDBTitle: structure of formylglycinamide synthetase
|
|
|
| 28 | d1clib1 |
|
not modelled |
100.0 |
21 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 29 | d1clia1 |
|
not modelled |
100.0 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 30 | d2zaua1 |
|
not modelled |
100.0 |
15 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 31 | d3c9ua2 |
|
not modelled |
100.0 |
34 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 32 | d2zoda1 |
|
not modelled |
100.0 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 33 | c3mdoB_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:putative phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a putative phosphoribosylformylglycinamidine2 cyclo-ligase (bdi_2101) from parabacteroides distasonis atcc 8503 at3 1.91 a resolution
|
|
|
| 34 | d2z1ea1 |
|
not modelled |
100.0 |
24 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 35 | d1vk3a2 |
|
not modelled |
99.9 |
14 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 36 | d2zoda2 |
|
not modelled |
99.9 |
15 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 37 | c3kizA_ |
|
not modelled |
99.9 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of putative phosphoribosylformylglycinamidine cyclo-2 ligase (yp_676759.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
|
|
|
| 38 | d2z1ea2 |
|
not modelled |
99.9 |
25 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 39 | d1vk3a1 |
|
not modelled |
99.9 |
23 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 40 | d1clia2 |
|
not modelled |
99.9 |
17 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 41 | d1vk3a3 |
|
not modelled |
99.8 |
13 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 42 | d1t3ta4 |
|
not modelled |
99.6 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 43 | d1t3ta6 |
|
not modelled |
99.6 |
14 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 44 | d1t3ta5 |
|
not modelled |
99.2 |
16 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 45 | d1t3ta7 |
|
not modelled |
99.2 |
23 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 46 | c3fokH_ |
|
not modelled |
86.2 |
17 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein cgl0159;
PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
|
|
|
| 47 | c3jrkG_ |
|
not modelled |
84.1 |
15 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
|
|
| 48 | d1to3a_ |
|
not modelled |
76.9 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 49 | c3qi7A_ |
|
not modelled |
73.1 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
|
|
|
| 50 | c3dzvB_ |
|
not modelled |
42.9 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:4-methyl-5-(beta-hydroxyethyl)thiazole kinase;
PDBTitle: crystal structure of 4-methyl-5-(beta-hydroxyethyl)thiazole kinase2 (np_816404.1) from enterococcus faecalis v583 at 2.57 a resolution
|
|
|
| 51 | c2qjhH_ |
|
not modelled |
42.6 |
19 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|
|
|
| 52 | c4ru1C_ |
|
not modelled |
41.2 |
5 |
PDB header:transport protein
Chain: C: PDB Molecule:monosaccharide abc transporter substrate-binding protein,
PDBTitle: crystal structure of carbohydrate transporter acei_1806 from2 acidothermus cellulolyticus 11b, target efi-510965, in complex with3 myo-inositol
|
|
|
| 53 | d2csua1 |
|
not modelled |
38.6 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 54 | c4mozC_ |
|
not modelled |
34.1 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: fructose-bisphosphate aldolase from slackia heliotrinireducens dsm2 20476
|
|
|
| 55 | c2hqbA_ |
|
not modelled |
33.7 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional activator of comk gene;
PDBTitle: crystal structure of a transcriptional activator of comk2 gene from bacillus halodurans
|
|
|
| 56 | c5jxpA_ |
|
not modelled |
32.7 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:asp/glu-specific dipeptidyl-peptidase;
PDBTitle: crystal structure of porphyromonas endodontalis dpp11 in alternate2 conformation
|
|
|
| 57 | c5jxfA_ |
|
not modelled |
29.2 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:asp/glu-specific dipeptidyl-peptidase;
PDBTitle: crystal structure of flavobacterium psychrophilum dpp11 in complex2 with dipeptide arg-asp
|
|
|
| 58 | c5kdmD_ |
|
not modelled |
28.2 |
15 |
PDB header:chaperone / dna binding protein
Chain: D: PDB Molecule:major tegument protein;
PDBTitle: crystal structure of ebv tegument protein bnrf1 in complex with2 histone chaperone daxx and histones h3.3-h4
|
|
|
| 59 | c3gndC_ |
|
not modelled |
24.6 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
|
|
| 60 | d1fnda1 |
|
not modelled |
23.3 |
25 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 61 | d1wf8a1 |
|
not modelled |
22.5 |
11 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 62 | c3g85A_ |
|
not modelled |
21.4 |
5 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (laci family);
PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
|
|
|
| 63 | d1v8aa_ |
|
not modelled |
21.0 |
29 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Thiamin biosynthesis kinases |
|
|
| 64 | c3eggC_ |
|
not modelled |
20.7 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:spinophilin;
PDBTitle: crystal structure of a complex between protein phosphatase 1 alpha2 (pp1) and the pp1 binding and pdz domains of spinophilin
|
|
|
| 65 | c3lukB_ |
|
not modelled |
20.5 |
9 |
PDB header:rna binding protein
Chain: B: PDB Molecule:protein argonaute-2;
PDBTitle: crystal structure of mid domain from hago2
|
|
|
| 66 | d1gawa1 |
|
not modelled |
20.1 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 67 | c5mvrA_ |
|
not modelled |
19.9 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:trna threonylcarbamoyladenosine biosynthesis protein tsae;
PDBTitle: crystal structure of bacillus subtilus ydib
|
|
|
| 68 | d2bmwa1 |
|
not modelled |
19.8 |
22 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 69 | c5braA_ |
|
not modelled |
19.4 |
14 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:putative periplasmic binding protein with substrate ribose;
PDBTitle: crystal structure of a putative periplasmic solute binding protein2 (ipr025997) from ochrobactrum anthropi atcc49188 (oant_2843, target3 efi-511085)
|
|
|
| 70 | d1htwa_ |
|
not modelled |
19.4 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:YjeE-like |
|
|
| 71 | c4y01B_ |
|
not modelled |
18.9 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase s46;
PDBTitle: crystal structure of dipeptidyl peptidase 11 (dpp11) from2 porphyromonas gingivalis
|
|
|
| 72 | d1ep3b1 |
|
not modelled |
18.7 |
32 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 73 | c2jreA_ |
|
not modelled |
18.3 |
16 |
PDB header:de novo protein
Chain: A: PDB Molecule:c60-1 pdz domain peptide;
PDBTitle: c60-1, a pdz domain designed using statistical coupling2 analysis
|
|
|
| 74 | c3rotA_ |
|
not modelled |
18.1 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:abc sugar transporter, periplasmic sugar binding protein;
PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
|
|
|
| 75 | d2byga1 |
|
not modelled |
18.0 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 76 | c5f67A_ |
|
not modelled |
17.9 |
23 |
PDB header:protein binding
Chain: A: PDB Molecule:inactivation-no-after-potential d protein;
PDBTitle: an exquisitely specific pdz/target recognition revealed by the2 structure of inad pdz3 in complex with trp channel tail
|
|
|
| 77 | c6ndiB_ |
|
not modelled |
17.7 |
15 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: 2.60 angstrom resolution crystal structure of periplasmic binding and2 sugar binding domain of laci family protein from klebsiella3 pneumoniae.
|
|
|
| 78 | c3wolB_ |
|
not modelled |
17.5 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:dipeptidyl aminopeptidase bii;
PDBTitle: crystal structure of the dap bii dipeptide complex i
|
|
|
| 79 | d1jyea_ |
|
not modelled |
17.2 |
21 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 80 | c1jyeA_ |
|
not modelled |
17.2 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:lactose operon repressor;
PDBTitle: structure of a dimeric lac repressor with c-terminal deletion and k84l2 substitution
|
|
|
| 81 | c6dspB_ |
|
not modelled |
17.1 |
17 |
PDB header:signaling protein
Chain: B: PDB Molecule:autoinducer 2-binding protein lsrb;
PDBTitle: lsrb from clostridium saccharobutylicum in complex with ai-2
|
|
|
| 82 | c3d02A_ |
|
not modelled |
17.1 |
3 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative laci-type transcriptional regulator;
PDBTitle: crystal structure of periplasmic sugar-binding protein2 (yp_001338366.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 1.30 a resolution
|
|
|
| 83 | d2f0aa1 |
|
not modelled |
16.7 |
25 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 84 | d1ujda_ |
|
not modelled |
16.6 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 85 | c3qz6A_ |
|
not modelled |
16.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
|
|
| 86 | d1oi7a1 |
|
not modelled |
16.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 87 | d1x5qa1 |
|
not modelled |
15.8 |
23 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 88 | c6fpeE_ |
|
not modelled |
15.6 |
30 |
PDB header:rna binding protein
Chain: E: PDB Molecule:atpase yjee, predicted to have essential role in cell wall
PDBTitle: bacterial protein complex
|
|
|
| 89 | c6hyhA_ |
|
not modelled |
15.4 |
17 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of msmeg_1712 from mycobacterium smegmatis in2 complex with beta-d-fucofuranose
|
|
|
| 90 | c3gbvB_ |
|
not modelled |
15.3 |
9 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative laci-family transcriptional regulator;
PDBTitle: crystal structure of a putative laci transcriptional regulator from2 bacteroides fragilis
|
|
|
| 91 | d1tjya_ |
|
not modelled |
14.3 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 92 | d1wfga_ |
|
not modelled |
13.9 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 93 | c3dbiA_ |
|
not modelled |
13.8 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:sugar-binding transcriptional regulator, laci family;
PDBTitle: crystal structure of sugar-binding transcriptional regulator (laci2 family) from escherichia coli complexed with phosphate
|
|
|
| 94 | c4ywkA_ |
|
not modelled |
13.8 |
31 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division control protein 21;
PDBTitle: pyrococcus furiosus mcm n-terminal domain with zinc-binding subdomain2 b deleted
|
|
|
| 95 | c4wzzA_ |
|
not modelled |
13.7 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:putative sugar abc transporter, substrate-binding protein;
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from clostridium phytofermentas (cphy_0583, target efi-3 511148) with bound l-rhamnose
|
|
|
| 96 | c5hqjA_ |
|
not modelled |
13.6 |
15 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of abc transporter solute binding protein b1g1h72 from burkholderia graminis c4d1m, target efi-511179, in complex with3 d-arabinose
|
|
|
| 97 | d1whaa_ |
|
not modelled |
13.6 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 98 | c3d8uA_ |
|
not modelled |
13.5 |
6 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
|
|
| 99 | c4kzkA_ |
|
not modelled |
13.5 |
12 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:l-arabinose abc transporter, periplasmic l-arabinose-
PDBTitle: the structure of the periplasmic l-arabinose binding protein from2 burkholderia thailandensis
|
|
|