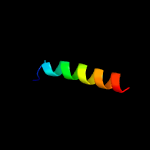| 1 |
|
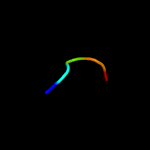
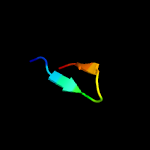
PDB 3a0h chain J
Region: 9 - 39
Aligned: 31
Modelled: 31
Confidence: 22.6%
Identity: 16%
PDB header:electron transport
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 2 |
|
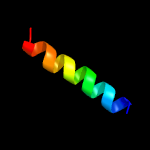
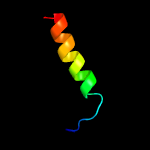
PDB 2axt chain J domain 1
Region: 9 - 39
Aligned: 31
Modelled: 31
Confidence: 22.6%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2
| 3 |
|
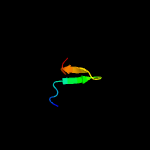
PDB 1f2q chain A domain 1
Region: 80 - 143
Aligned: 47
Modelled: 64
Confidence: 15.8%
Identity: 13%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 4 |
|
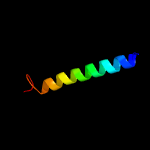
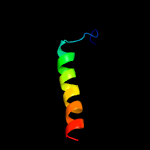
PDB 1su3 chain A domain 1
Region: 98 - 102
Aligned: 5
Modelled: 5
Confidence: 14.0%
Identity: 60%
Fold: PGBD-like
Superfamily: PGBD-like
Family: MMP N-terminal domain
Phyre2
| 5 |
|
PDB 1slm chain A domain 1
Region: 98 - 102
Aligned: 5
Modelled: 5
Confidence: 13.6%
Identity: 60%
Fold: PGBD-like
Superfamily: PGBD-like
Family: MMP N-terminal domain
Phyre2
| 6 |
|
PDB 1eak chain A domain 1
Region: 98 - 102
Aligned: 5
Modelled: 5
Confidence: 12.7%
Identity: 60%
Fold: PGBD-like
Superfamily: PGBD-like
Family: MMP N-terminal domain
Phyre2
| 7 |
|
PDB 6f0k chain A
Region: 8 - 42
Aligned: 35
Modelled: 35
Confidence: 11.9%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:cytochrome c family protein;
PDBTitle: alternative complex iii
Phyre2
| 8 |
|
PDB 1l6j chain A domain 1
Region: 98 - 102
Aligned: 5
Modelled: 5
Confidence: 11.6%
Identity: 60%
Fold: PGBD-like
Superfamily: PGBD-like
Family: MMP N-terminal domain
Phyre2
| 9 |
|
PDB 4xtr chain G
Region: 10 - 29
Aligned: 20
Modelled: 20
Confidence: 9.2%
Identity: 15%
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:pep12p;
PDBTitle: structure of get3 bound to the transmembrane domain of pep12
Phyre2
| 10 |
|
PDB 5eur chain C
Region: 90 - 107
Aligned: 18
Modelled: 18
Confidence: 8.6%
Identity: 11%
PDB header:unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: hypothetical protein sf216 from shigella flexneri 5a m90t
Phyre2
| 11 |
|
PDB 2l2t chain A
Region: 10 - 31
Aligned: 22
Modelled: 22
Confidence: 8.3%
Identity: 36%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 12 |
|
PDB 2lcx chain B
Region: 10 - 31
Aligned: 22
Modelled: 22
Confidence: 8.3%
Identity: 36%
PDB header:transferase
Chain: B: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: spatial structure of the erbb4 dimeric tm domain
Phyre2
| 13 |
|
PDB 3jcu chain J
Region: 9 - 39
Aligned: 31
Modelled: 31
Confidence: 8.1%
Identity: 10%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
Phyre2
| 14 |
|
PDB 3kpa chain B
Region: 117 - 138
Aligned: 18
Modelled: 22
Confidence: 7.6%
Identity: 28%
PDB header:ligase
Chain: B: PDB Molecule:probable ubiquitin fold modifier conjugating enzyme;
PDBTitle: ubiquitin fold modifier conjugating enzyme from leishmania major2 (probable)
Phyre2
| 15 |
|
PDB 6eyv chain B
Region: 112 - 122
Aligned: 11
Modelled: 11
Confidence: 7.3%
Identity: 36%
PDB header:oxidoreductase
Chain: B: PDB Molecule:pvdp;
PDBTitle: crystal structure of the pyoverdine maturation protein pvdp in complex2 with the mock substrates l-tyrosine and zinc.
Phyre2
| 16 |
|
PDB 1ic9 chain A
Region: 94 - 99
Aligned: 6
Modelled: 6
Confidence: 7.2%
Identity: 50%
PDB header:de novo protein
Chain: A: PDB Molecule:th10aox;
PDBTitle: nmr solution structure of the designed beta-sheet mini-2 protein th10aox
Phyre2
| 17 |
|
PDB 2knc chain B
Region: 8 - 30
Aligned: 23
Modelled: 23
Confidence: 6.9%
Identity: 9%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 18 |
|
PDB 2m20 chain B
Region: 10 - 31
Aligned: 22
Modelled: 22
Confidence: 6.4%
Identity: 14%
PDB header:signaling protein
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: egfr transmembrane - juxtamembrane (tm-jm) segment in bicelles: md2 guided nmr refined structure.
Phyre2
| 19 |
|
PDB 2l8s chain A
Region: 3 - 30
Aligned: 28
Modelled: 28
Confidence: 5.8%
Identity: 14%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-1;
PDBTitle: solution nmr structure of transmembrane and cytosolic regions of2 integrin alpha1 in detergent micelles
Phyre2
| 20 |
|
PDB 1ugn chain A domain 1
Region: 90 - 139
Aligned: 49
Modelled: 50
Confidence: 5.7%
Identity: 24%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: I set domains
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|