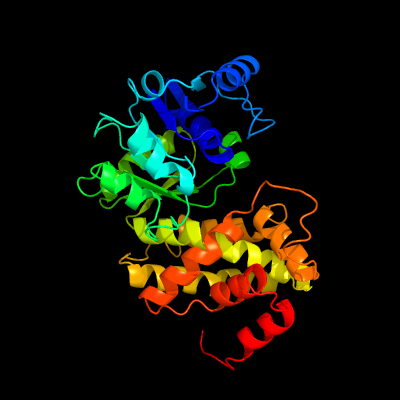
| 1 | c1m67A_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of leishmania mexicana gpdh complexed with inhibitor2 2-bromo-6-hydroxy-purine
|
|
|
|
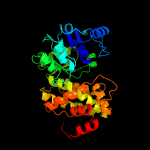
| 2 | c3k96B_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
|
|
|
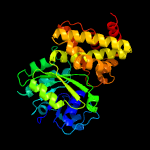
| 3 | c1wpqB_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad+],
PDBTitle: ternary complex of glycerol 3-phosphate dehydrogenase 12 with nad and dihydroxyactone
|
|
|
|
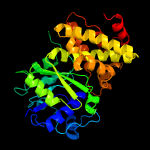
| 4 | c1yj8C_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: initial structural analysis of plasmodium falciparum glycerol-3-2 phosphate dehydrogenase
|
|
|
|
| 5 | c1z82A_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glycerol-3-phosphate dehydrogenase (tm0378) from2 thermotoga maritima at 2.00 a resolution
|
|
|
|
| 6 | c4fgwA_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(+)] 1;
PDBTitle: structure of glycerol-3-phosphate dehydrogenase, gpd1, from2 sacharomyces cerevisiae
|
|
|
|
| 7 | c1txgA_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: structure of glycerol-3-phosphate dehydrogenase from archaeoglobus2 fulgidus
|
|
|
|
| 8 | c2ew2B_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase, putative;
PDBTitle: crystal structure of the putative 2-dehydropantoate 2-reductase from2 enterococcus faecalis
|
|
|
|
| 9 | c3hn2A_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from geobacter2 metallireducens gs-15
|
|
|
|
| 10 | c1bg6A_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;
PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
|
|
|
|
| 11 | c3hwrA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of pane/apba family ketopantoate reductase2 (yp_299159.1) from ralstonia eutropha jmp134 at 2.15 a resolution
|
|
|
|
| 12 | c5zikC_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:probable 2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of ketopantoate reductase from pseudomonas2 aeruginosa
|
|
|
|
| 13 | c2qytA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from porphyromonas2 gingivalis w83
|
|
|
|
| 14 | c5ayvB_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of archaeal ketopantoate reductase complexed with2 coenzyme a and 2-oxopantoate
|
|
|
|
| 15 | c4ol9A_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of putative 2-dehydropantoate 2-reductase pane from2 mycobacterium tuberculosis complexed with nadp and oxamate
|
|
|
|
| 16 | c3ghyA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ketopantoate reductase protein;
PDBTitle: crystal structure of a putative ketopantoate reductase from ralstonia2 solanacearum molk2
|
|
|
|
| 17 | c2ofpB_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketopantoate reductase;
PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
|
|
|
|
| 18 | c3wfjD_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: the complex structure of d-mandelate dehydrogenase with nadh
|
|
|
|
| 19 | c1ks9A_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: ketopantoate reductase from escherichia coli
|
|
|
|
| 20 | c3egoB_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable 2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of probable 2-dehydropantoate 2-reductase pane from2 bacillus subtilis
|
|
|
|
| 21 | c3g17H_ |
|
not modelled |
100.0 |
14 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:similar to 2-dehydropantoate 2-reductase;
PDBTitle: structure of putative 2-dehydropantoate 2-reductase from2 staphylococcus aureus
|
|
|
| 22 | c3i83B_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of 2-dehydropantoate 2-reductase from methylococcus2 capsulatus
|
|
|
| 23 | c2y0dB_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: bcec mutation y10k
|
|
|
| 24 | c1mv8A_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gdp-mannose 6-dehydrogenase;
PDBTitle: 1.55 a crystal structure of a ternary complex of gdp-mannose2 dehydrogenase from psuedomonas aeruginosa
|
|
|
| 25 | c3gg2B_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sugar dehydrogenase, udp-glucose/gdp-mannose dehydrogenase
PDBTitle: crystal structure of udp-glucose 6-dehydrogenase from porphyromonas2 gingivalis bound to product udp-glucuronate
|
|
|
| 26 | c3vtfA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of a udp-glucose dehydrogenase from the hyperthermophilic2 archaeon pyrobaculum islandicum
|
|
|
| 27 | c4a7pA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: se-met derivatized ugdg, udp-glucose dehydrogenase from sphingomonas2 elodea
|
|
|
| 28 | c3qhaB_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
|
|
| 29 | c4wb1B_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cals8;
PDBTitle: crystal structure of cals8 from micromonospora echinospora (p294s2 mutant)
|
|
|
| 30 | c3w6uA_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, nad-binding protein;
PDBTitle: crystal structure of nadp bound l-serine 3-dehydrogenase from2 hyperthermophilic archaeon pyrobaculum calidifontis
|
|
|
| 31 | c3cumA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of a possible 3-hydroxyisobutyrate dehydrogenase2 from pseudomonas aeruginosa pao1
|
|
|
| 32 | c3prjB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: role of packing defects in the evolution of allostery and induced fit2 in human udp-glucose dehydrogenase.
|
|
|
| 33 | c2q3eH_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
|
|
|
| 34 | c5y8mA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxyisobutyrate dehydrogenase;
PDBTitle: mycobacterium tuberculosis 3-hydroxyisobutyrate dehydrogenase2 (mthibadh) + nad + (r)-3-hydroxyisobutyrate (r-hiba)
|
|
|
| 35 | c1vpdA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
|
|
| 36 | c2p4qA_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating 1;
PDBTitle: crystal structure analysis of gnd1 in saccharomyces cerevisiae
|
|
|
| 37 | c2cvzD_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: structure of hydroxyisobutyrate dehydrogenase from thermus2 thermophilus hb8
|
|
|
| 38 | c1yb4A_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronic semialdehyde reductase;
PDBTitle: crystal structure of the tartronic semialdehyde reductase from2 salmonella typhimurium lt2
|
|
|
| 39 | c2o3jC_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of caenorhabditis elegans udp-glucose dehydrogenase
|
|
|
| 40 | c4e21B_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase (decarboxylating);
PDBTitle: the crystal structure of 6-phosphogluconate dehydrogenase from2 geobacter metallireducens
|
|
|
| 41 | c3c7cB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
|
|
| 42 | c4dllB_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-hydroxy-3-oxopropionate reductase;
PDBTitle: crystal structure of a 2-hydroxy-3-oxopropionate reductase from2 polaromonas sp. js666
|
|
|
| 43 | c3g0oA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of 3-hydroxyisobutyrate dehydrogenase2 (ygbj) from salmonella typhimurium
|
|
|
| 44 | c1pgqA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase (choh(d)-nadp+(a))
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: crystallographic study of coenzyme, coenzyme analogue and substrate2 binding in 6-phosphogluconate dehydrogenase: implications for nadp3 specificity and the enzyme mechanism
|
|
|
| 45 | c1pgjA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
|
|
|
| 46 | c4r16A_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:418aa long hypothetical udp-n-acetyl-d-mannosaminuronic
PDBTitle: structure of udp-d-mannac dehdrogeanse from pyrococcus horikoshii
|
|
|
| 47 | c4gbjB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase nad-binding;
PDBTitle: crystal structure of nad-binding 6-phosphogluconate dehydrogenase from2 dyadobacter fermentans
|
|
|
| 48 | c3ckyA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-hydroxymethyl glutarate dehydrogenase;
PDBTitle: structural and kinetic properties of a beta-hydroxyacid dehydrogenase2 involved in nicotinate fermentation
|
|
|
| 49 | c2iz1C_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
|
|
| 50 | c5je8A_ |
|
not modelled |
99.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: the crystal structure of bacillus cereus 3-hydroxyisobutyrate2 dehydrogenase in complex with nad
|
|
|
| 51 | c5a9tA_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative dehydrogenase;
PDBTitle: imine reductase from amycolatopsis orientalis in complex2 with (r)-methyltetrahydroisoquinoline
|
|
|
| 52 | c3fwnB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
|
|
|
| 53 | c4oqzA_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase yfjr;
PDBTitle: streptomyces aurantiacus imine reductase
|
|
|
| 54 | c2gf2B_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
|
|
|
| 55 | c6fqzB_ |
|
not modelled |
99.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: plasmodium falciparum 6-phosphogluconate dehydrogenase in its apo2 form, in complex with its cofactor nadp+ and in complex with its3 substrate 6-phosphogluconate
|
|
|
| 56 | c5ocmA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad_gly3p_dh, nad-dependent glycerol-3-phosphate
PDBTitle: imine reductase from streptosporangium roseum in complex with nadp+2 and 2,2,2-trifluoroacetophenone hydrate
|
|
|
| 57 | c3g79A_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ndp-n-acetyl-d-galactosaminuronic acid dehydrogenase;
PDBTitle: crystal structure of ndp-n-acetyl-d-galactosaminuronic acid2 dehydrogenase from methanosarcina mazei go1
|
|
|
| 58 | c4kqxB_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: mutant slackia exigua kari ddv in complex with nad and an inhibitor
|
|
|
| 59 | c6aqjB_ |
|
not modelled |
99.9 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:ketol-acid reductoisomerase (nadp(+));
PDBTitle: crystal structures of staphylococcus aureus ketol-acid2 reductoisomerase in complex with two transition state analogs that3 have biocidal activity.
|
|
|
| 60 | c3plnA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
|
|
|
| 61 | c3l6dB_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
|
|
| 62 | d1n1ea2 |
|
not modelled |
99.9 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 63 | c3dojA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase-like protein;
PDBTitle: structure of glyoxylate reductase 1 from arabidopsis (atglyr1)
|
|
|
| 64 | c5g6sD_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:imine reductase;
PDBTitle: imine reductase from aspergillus oryzae in complex with nadp(h) and2 (r)-rasagiline
|
|
|
| 65 | c5u5gC_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase;
PDBTitle: psf3 in complex with nadp+ and 2-opp
|
|
|
| 66 | c4d3fB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:imine reductase;
PDBTitle: bcsired from bacillus cereus in complex with nadph
|
|
|
| 67 | c3zhbC_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:r-imine reductase;
PDBTitle: r-imine reductase from streptomyces kanamyceticus in2 complex with nadp.
|
|
|
| 68 | c3ojlA_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cap5o;
PDBTitle: native structure of the udp-n-acetyl-mannosamine dehydrogenase cap5o2 from staphylococcus aureus
|
|
|
| 69 | c2uyyD_ |
|
not modelled |
99.9 |
17 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
|
|
| 70 | c4oqyA_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(s)-imine reductase;
PDBTitle: streptomyces sp. gf3546 imine reductase
|
|
|
| 71 | c4d3sA_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:imine reductase;
PDBTitle: imine reductase from nocardiopsis halophila
|
|
|
| 72 | c1dliA_ |
|
not modelled |
99.9 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose dehydrogenase;
PDBTitle: the first structure of udp-glucose dehydrogenase (udpgdh) reveals the2 catalytic residues necessary for the two-fold oxidation
|
|
|
| 73 | c3pduF_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:3-hydroxyisobutyrate dehydrogenase family protein;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter sulfurreducens in complex with nadp+
|
|
|
| 74 | c6grlA_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate
PDBTitle: structure of imine reductase (apo form) at 1.6 a resolution from2 saccharomonospora xinjiangensis
|
|
|
| 75 | c5ojlA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:imine reductase;
PDBTitle: imine reductase from aspergillus terreus in complex with nadph4 and2 dibenz[c,e]azepine
|
|
|
| 76 | c4xdzB_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: holo structure of ketol-acid reductoisomerase from ignisphaera2 aggregans
|
|
|
| 77 | c4edfC_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: dimeric hugdh, k94e
|
|
|
| 78 | d1txga2 |
|
not modelled |
99.9 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 79 | c6c4nB_ |
|
not modelled |
99.9 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pseudopaline dehydrogenase;
PDBTitle: pseudopaline dehydrogenase (paodh) - nadp+ bound
|
|
|
| 80 | c6c4lC_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:yersinopine dehydrogenase;
PDBTitle: yersinopine dehydrogenase (ypodh) - apo
|
|
|
| 81 | c3pefA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:6-phosphogluconate dehydrogenase, nad-binding;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter metallireducens in complex with nadp+
|
|
|
| 82 | c4ezbA_ |
|
not modelled |
99.9 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of the conserved hypothetical protein from2 sinorhizobium meliloti 1021
|
|
|
| 83 | c4xdyB_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: structure of nadh-preferring ketol-acid reductoisomerase from an2 uncultured archean
|
|
|
| 84 | c3qsgA_ |
|
not modelled |
99.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nad-binding phosphogluconate dehydrogenase-like protein;
PDBTitle: crystal structure of nad-binding phosphogluconate dehydrogenase-like2 protein from alicyclobacillus acidocaldarius
|
|
|
| 85 | c5yeqB_ |
|
not modelled |
99.9 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:ketol-acid reductoisomerase (nadp(+));
PDBTitle: the structure of sac-kari protein
|
|
|
| 86 | d1n1ea1 |
|
not modelled |
99.9 |
39 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:Glycerol-3-phosphate dehydrogenase |
|
|
| 87 | c6c4rA_ |
|
not modelled |
99.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:staphylopine dehydrogenase;
PDBTitle: staphylopine dehydrogenase (saodh) - apo
|
|
|
| 88 | c5t57A_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:semialdehyde dehydrogenase nad-binding protein;
PDBTitle: crystal structure of a semialdehyde dehydrogenase nad-binding protein2 from cupriavidus necator in complex with calcium and nad
|
|
|
| 89 | c6c4jA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: ligand bound full length hugdh with a104l substitution
|
|
|
| 90 | c3d1lB_ |
|
not modelled |
99.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadp oxidoreductase bf3122;
PDBTitle: crystal structure of putative nadp oxidoreductase bf3122 from2 bacteroides fragilis
|
|
|
| 91 | d1bg6a2 |
|
not modelled |
99.8 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 92 | c2izzE_ |
|
not modelled |
99.8 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyrroline-5-carboxylate reductase 1;
PDBTitle: crystal structure of human pyrroline-5-carboxylate reductase
|
|
|
| 93 | c4e12A_ |
|
not modelled |
99.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diketoreductase;
PDBTitle: substrate-directed dual catalysis of dicarbonyl compounds by2 diketoreductase
|
|
|
| 94 | d1mv8a2 |
|
not modelled |
99.8 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 95 | c4j0eB_ |
|
not modelled |
99.8 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable 3-hydroxyacyl-coa dehydrogenase f54c8.1;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase from2 caenorhadbitis elegans in p1 space group
|
|
|
| 96 | c2graA_ |
|
not modelled |
99.8 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase 1;
PDBTitle: crystal structure of human pyrroline-5-carboxylate reductase complexed2 with nadp
|
|
|
| 97 | c5bseF_ |
|
not modelled |
99.8 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: crystal structure of medicago truncatula (delta)1-pyrroline-5-2 carboxylate reductase (mtp5cr)
|
|
|
| 98 | c4ypoB_ |
|
not modelled |
99.8 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of mycobacterium tuberculosis ketol-acid2 reductoisomerase in complex with mg2+
|
|
|
| 99 | d1txga1 |
|
not modelled |
99.8 |
36 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:Glycerol-3-phosphate dehydrogenase |
|
|
| 100 | c2ep9A_ |
|
not modelled |
99.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-gulonate 3-dehydrogenase;
PDBTitle: crystal structure of the rabbit l-gulonate 3-dehydrogenase2 (nadh form)
|
|
|
| 101 | c1i36A_ |
|
not modelled |
99.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function reveals2 structural similarity with 3-hydroxyacid dehydrogenases
|
|
|
| 102 | c3triB_ |
|
not modelled |
99.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
|
|
|
| 103 | c2ahrB_ |
|
not modelled |
99.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative pyrroline carboxylate reductase;
PDBTitle: crystal structures of 1-pyrroline-5-carboxylate reductase from human2 pathogen streptococcus pyogenes
|
|
|
| 104 | c4om8B_ |
|
not modelled |
99.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxybutyryl-coa dehydrogenase;
PDBTitle: crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic2 acid (fhmpc) 5-dehydrogenase, an nad+ dependent dismutase.
|
|
|
| 105 | d2pgda2 |
|
not modelled |
99.8 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 106 | d1ks9a2 |
|
not modelled |
99.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 107 | c1np3B_ |
|
not modelled |
99.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
|
|
|
| 108 | c5n2iC_ |
|
not modelled |
99.8 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:reduced coenzyme f420:nadp oxidoreductase;
PDBTitle: f420:nadph oxidoreductase from thermobifida fusca with nadp+ bound
|
|
|
| 109 | c3dttA_ |
|
not modelled |
99.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp oxidoreductase;
PDBTitle: crystal structure of a putative f420 dependent nadp-reductase2 (arth_0613) from arthrobacter sp. fb24 at 1.70 a resolution
|
|
|
| 110 | c3k6jA_ |
|
not modelled |
99.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein f01g10.3, confirmed by transcript evidence;
PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
|
|
|
| 111 | c3rqsB_ |
|
not modelled |
99.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:hydroxyacyl-coenzyme a dehydrogenase, mitochondrial;
PDBTitle: crystal structure of human l-3- hydroxyacyl-coa dehydrogenase2 (ec1.1.1.35) from mitochondria at the resolution 2.0 a, northeast3 structural genomics consortium target hr487, mitochondrial protein4 partnership
|
|
|
| 112 | c3mogA_ |
|
not modelled |
99.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable 3-hydroxybutyryl-coa dehydrogenase;
PDBTitle: crystal structure of 3-hydroxybutyryl-coa dehydrogenase from2 escherichia coli k12 substr. mg1655
|
|
|
| 113 | c1m75B_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyacyl-coa dehydrogenase;
PDBTitle: crystal structure of the n208s mutant of l-3-hydroxyacyl-coa2 dehydrogenase in complex with nad and acetoacetyl-coa
|
|
|
| 114 | c2f1kD_ |
|
not modelled |
99.7 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
|
|
| 115 | d1vpda2 |
|
not modelled |
99.7 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 116 | c6hrdD_ |
|
not modelled |
99.7 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-hydroxybutyryl-coa dehydrogenase;
PDBTitle: crystal structure of m. tuberculosis fadb2 (rv0468)
|
|
|
| 117 | c4kueA_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxybutyryl-coa dehydrogenase;
PDBTitle: crystal structure of 3-hydroxybutylryl-coa dehydrogenase from2 clostridium butyricum
|
|
|
| 118 | d1jaya_ |
|
not modelled |
99.7 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 119 | c3gt0A_ |
|
not modelled |
99.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: crystal structure of pyrroline 5-carboxylate reductase from bacillus2 cereus. northeast structural genomics consortium target bcr38b
|
|
|
| 120 | d2f1ka2 |
|
not modelled |
99.7 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|