| 1 | c1xdoB_
|
|
 |
100.0 |
33 |
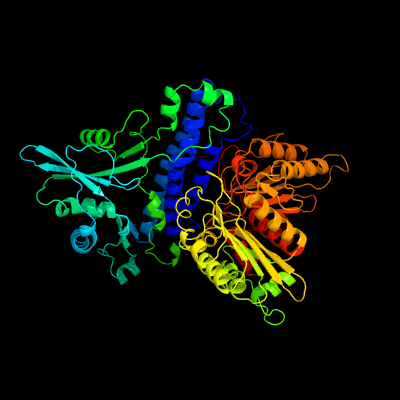
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
|
|
|
| 2 | c2o8rA_
|
|
 |
100.0 |
34 |
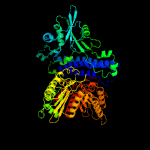
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
|
|
|
|
| 3 | d2o8ra3
|
|
 |
100.0 |
36 |
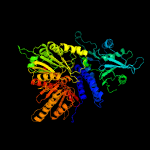
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
|
| 4 | d2o8ra4
|
|
 |
100.0 |
33 |
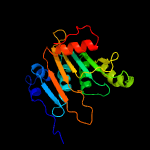
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
|
| 5 | d1xdpa4
|
|
 |
100.0 |
32 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
|
| 6 | d1xdpa2
|
|
 |
100.0 |
22 |
Fold:PHP14-like
Superfamily:PHP14-like
Family:PPK middle domain-like |
|
|
|
| 7 | d1xdpa3
|
|
 |
100.0 |
43 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
|
| 8 | d2o8ra2
|
|
 |
100.0 |
27 |
Fold:PHP14-like
Superfamily:PHP14-like
Family:PPK middle domain-like |
|
|
|
| 9 | d1xdpa1
|
|
 |
100.0 |
40 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
|
|
|
| 10 | d2o8ra1
|
|
 |
100.0 |
44 |
Fold:Spectrin repeat-like
Superfamily:PPK N-terminal domain-like
Family:PPK N-terminal domain-like |
|
|
|
| 11 | c3hsiC_
|
|
 |
100.0 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:phosphatidylserine synthase;
PDBTitle: crystal structure of phosphatidylserine synthase haemophilus2 influenzae rd kw20
|
|
|
|
| 12 | c1v0sA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:phospholipase d;
PDBTitle: uninhibited form of phospholipase d from streptomyces sp.2 strain pmf
|
|
|
|
| 13 | c4ggkA_
|
|
 |
99.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:mitochondrial cardiolipin hydrolase;
PDBTitle: crystal structure of zucchini from mouse (mzuc / pld6 / mitopld) bound2 to tungstate
|
|
|
|
| 14 | d1byra_
|
|
 |
99.5 |
16 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Nuclease |
|
|
|
| 15 | c6ehiI_
|
|
 |
99.5 |
10 |
PDB header:dna binding protein
Chain: I: PDB Molecule:nuclease nuct;
PDBTitle: nuct from helicobacter pylori
|
|
|
|
| 16 | c4gelA_
|
|
 |
99.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:mitochondrial cardiolipin hydrolase;
PDBTitle: crystal structure of zucchini
|
|
|
|
| 17 | c5lzkB_
|
|
 |
99.4 |
18 |
PDB header:structural genomics
Chain: B: PDB Molecule:protein fam83b;
PDBTitle: structure of the domain of unknown function duf1669 from human fam83b
|
|
|
|
| 18 | d1v0wa2
|
|
 |
99.3 |
27 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
|
|
|
| 19 | c4urjA_
|
|
 |
99.3 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:protein fam83a;
PDBTitle: crystal structure of human bj-tsa-9
|
|
|
|
| 20 | d1v0wa1
|
|
 |
99.2 |
18 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Phospholipase D |
|
|
|
| 21 | c4genA_ |
|
not modelled |
99.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:mitochondrial cardiolipin hydrolase;
PDBTitle: crystal structure of zucchini (monomer)
|
|
|
| 22 | c5bqtC_ |
|
not modelled |
97.7 |
9 |
PDB header:dna binding protein
Chain: C: PDB Molecule:putative hth-type transcriptional regulator trmbl2;
PDBTitle: structure of trmbl2, an archaeal chromatin protein, shows a novel mode2 of dna binding.
|
|
|
| 23 | c4rctB_ |
|
not modelled |
97.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:restriction endonuclease r.ngovii;
PDBTitle: crystal structure of r-protein of ngoavii restriction endonuclease
|
|
|
| 24 | c2c1lA_ |
|
not modelled |
94.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:restriction endonuclease;
PDBTitle: structure of the bfii restriction endonuclease
|
|
|
| 25 | c3qphA_ |
|
not modelled |
91.5 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:trmb, a global transcription regulator;
PDBTitle: the three-dimensional structure of trmb, a global transcriptional2 regulator of the hyperthermophilic archaeon pyrococcus furiosus in3 complex with sucrose
|
|
|
| 26 | c3ecsD_ |
|
not modelled |
90.9 |
29 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: crystal structure of human eif2b alpha
|
|
|
| 27 | d1vb5a_ |
|
not modelled |
90.3 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
|
|
| 28 | c6i7tB_ |
|
not modelled |
89.6 |
21 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: eif2b:eif2 complex
|
|
|
| 29 | c5b04B_ |
|
not modelled |
88.1 |
15 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 30 | d1zj8a2 |
|
not modelled |
87.8 |
22 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
|
|
| 31 | c4ldrA_ |
|
not modelled |
87.1 |
14 |
PDB header:isomerase, cell invasion
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: structure of the s283y mutant of mrdi
|
|
|
| 32 | c2yv2A_ |
|
not modelled |
86.8 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 aeropyrum pernix k1
|
|
|
| 33 | d1oy0a_ |
|
not modelled |
86.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 34 | c3r2jC_ |
|
not modelled |
85.2 |
31 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
|
|
| 35 | c5dboA_ |
|
not modelled |
84.8 |
23 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
|
|
|
| 36 | d1eucb1 |
|
not modelled |
83.5 |
20 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 37 | c2fpgA_ |
|
not modelled |
82.7 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa synthetase in2 complex with gdp
|
|
|
| 38 | d1jy1a1 |
|
not modelled |
82.0 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
|
|
| 39 | c3a11D_ |
|
not modelled |
81.7 |
21 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta subunit;
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
|
|
| 40 | c3gbcA_ |
|
not modelled |
81.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
|
|
| 41 | d1t9ka_ |
|
not modelled |
80.1 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
|
|
| 42 | d1qzqa1 |
|
not modelled |
79.7 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
|
|
| 43 | c6melA_ |
|
not modelled |
79.6 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:succinate--coa ligase subunit alpha;
PDBTitle: succinyl-coa synthase from campylobacter jejuni
|
|
|
| 44 | c5b04C_ |
|
not modelled |
79.4 |
25 |
PDB header:translation
Chain: C: PDB Molecule:probable translation initiation factor eif-2b subunit beta;
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 45 | d2nu7b1 |
|
not modelled |
79.4 |
21 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 46 | d1t5oa_ |
|
not modelled |
79.1 |
18 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
|
|
| 47 | c4zemB_ |
|
not modelled |
79.0 |
19 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif2b-like protein,
PDBTitle: crystal structure of eif2b beta from chaetomium thermophilum
|
|
|
| 48 | c6a34B_ |
|
not modelled |
78.8 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:putative methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate isomerase from2 pyrococcus horikoshii ot3 - form i
|
|
|
| 49 | c4zeoH_ |
|
not modelled |
78.1 |
23 |
PDB header:translation
Chain: H: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of eif2b delta from chaetomium thermophilum
|
|
|
| 50 | c5b04G_ |
|
not modelled |
78.0 |
23 |
PDB header:translation
Chain: G: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 51 | c1zj8B_ |
|
not modelled |
77.5 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable ferredoxin-dependent nitrite reductase nira;
PDBTitle: structure of mycobacterium tuberculosis nira protein
|
|
|
| 52 | d1oi7a2 |
|
not modelled |
76.8 |
20 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 53 | c6i3mD_ |
|
not modelled |
75.7 |
27 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit delta;
PDBTitle: eif2b:eif2 complex, phosphorylated on eif2 alpha serine 52.
|
|
|
| 54 | c6ezoD_ |
|
not modelled |
74.9 |
24 |
PDB header:membrane protein
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit beta;
PDBTitle: eukaryotic initiation factor eif2b in complex with isrib
|
|
|
| 55 | d2akja2 |
|
not modelled |
74.1 |
29 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
|
|
| 56 | c2b34C_ |
|
not modelled |
73.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
|
|
| 57 | c2yvkA_ |
|
not modelled |
72.2 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
|
|
| 58 | d1v59a2 |
|
not modelled |
72.0 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 59 | c6qg0G_ |
|
not modelled |
71.7 |
26 |
PDB header:translation
Chain: G: PDB Molecule:translation initiation factor eif-2b subunit delta;
PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model 1)
|
|
|
| 60 | c3b0nA_ |
|
not modelled |
70.1 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitrite reductase;
PDBTitle: q448k mutant of assimilatory nitrite reductase (nii3) from tobbaco2 leaf
|
|
|
| 61 | d2a0ua1 |
|
not modelled |
69.7 |
12 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
|
|
| 62 | c2h0rD_ |
|
not modelled |
68.1 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
|
|
| 63 | c4davA_ |
|
not modelled |
68.0 |
23 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:sugar fermentation stimulation protein homolog;
PDBTitle: the structure of pyrococcus furiosus sfsa in complex with dna
|
|
|
| 64 | c5n0sA_ |
|
not modelled |
67.5 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
| 65 | c1nopB_ |
|
not modelled |
67.4 |
11 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:tyrosyl-dna phosphodiesterase 1;
PDBTitle: crystal structure of human tyrosyl-dna phosphodiesterase2 (tdp1) in complex with vanadate, dna and a human3 topoisomerase i-derived peptide
|
|
|
| 66 | d1s4da_ |
|
not modelled |
66.9 |
19 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 67 | c5h92A_ |
|
not modelled |
66.7 |
14 |
PDB header:oxidoreductase/electron transport
Chain: A: PDB Molecule:sulfite reductase [ferredoxin], chloroplastic;
PDBTitle: crystal structure of the complex between maize sulfite reductase and2 ferredoxin in the form-3 crystal
|
|
|
| 68 | c2nu8D_ |
|
not modelled |
66.5 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
|
|
|
| 69 | c3mwdB_ |
|
not modelled |
66.4 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
|
|
| 70 | d2csua2 |
|
not modelled |
66.1 |
19 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 71 | c6g4qB_ |
|
not modelled |
65.9 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:succinate--coa ligase [adp-forming] subunit beta,
PDBTitle: structure of human adp-forming succinyl-coa ligase complex suclg1-2 sucla2
|
|
|
| 72 | c4dapA_ |
|
not modelled |
65.9 |
23 |
PDB header:dna binding protein
Chain: A: PDB Molecule:sugar fermentation stimulation protein a;
PDBTitle: the structure of escherichia coli sfsa
|
|
|
| 73 | c2yboA_ |
|
not modelled |
65.8 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
|
|
| 74 | d1o66a_ |
|
not modelled |
63.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 75 | c4yt2A_ |
|
not modelled |
62.8 |
42 |
PDB header:metal binding protein
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmd ii from methanocaldococcus jannaschii
|
|
|
| 76 | c3d3kD_ |
|
not modelled |
62.5 |
15 |
PDB header:protein binding
Chain: D: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of human edc3p
|
|
|
| 77 | c4v34A_ |
|
not modelled |
62.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:alanyl-trna-dependent l-alanyl- phophatidylglycerol
PDBTitle: the structure of a-pgs from pseudomonas aeruginosa (semet derivative)
|
|
|
| 78 | d1jy1a2 |
|
not modelled |
61.5 |
18 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
|
|
| 79 | c6huxA_ |
|
not modelled |
60.6 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmdii from methanocaldococcus jannaschii reconstitued with fe-2 guanylylpyridinol (fegp) cofactor and co-crystallized with methenyl-3 tetrahydromethanopterin at 2.5 a resolution
|
|
|
| 80 | c4xp7A_ |
|
not modelled |
60.2 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine(20) synthase [nad(p)+]-like;
PDBTitle: crystal structure of human trna dihydrouridine synthase 2
|
|
|
| 81 | c1yzvA_ |
|
not modelled |
59.5 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
|
|
| 82 | c1eucB_ |
|
not modelled |
59.1 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:succinyl-coa synthetase, beta chain;
PDBTitle: crystal structure of dephosphorylated pig heart, gtp-2 specific succinyl-coa synthetase
|
|
|
| 83 | d1vhna_ |
|
not modelled |
58.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 84 | c1nvmG_ |
|
not modelled |
57.6 |
10 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
| 85 | d1ebda2 |
|
not modelled |
55.4 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 86 | d1dxla2 |
|
not modelled |
53.6 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 87 | c4lrtC_ |
|
not modelled |
53.3 |
18 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
|
|
|
| 88 | d1euca2 |
|
not modelled |
53.2 |
21 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 89 | d1q32a2 |
|
not modelled |
52.5 |
20 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
|
|
| 90 | c6hxiD_ |
|
not modelled |
51.7 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:succinyl-coa ligase (adp-forming) subunit alpha;
PDBTitle: structure of atp citrate lyase from methanothrix soehngenii in complex2 with citrate and coenzyme a
|
|
|
| 91 | d1jpma1 |
|
not modelled |
51.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 92 | c2akjA_ |
|
not modelled |
51.5 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin--nitrite reductase, chloroplast;
PDBTitle: structure of spinach nitrite reductase
|
|
|
| 93 | d1j2ra_ |
|
not modelled |
50.9 |
12 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
|
|
| 94 | d1x9ga_ |
|
not modelled |
49.8 |
12 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
|
|
| 95 | d2nu7a1 |
|
not modelled |
49.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 96 | c3kwpA_ |
|
not modelled |
49.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
| 97 | d2nu7a2 |
|
not modelled |
49.2 |
24 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
|
|
| 98 | d1im5a_ |
|
not modelled |
48.6 |
11 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
|
|
| 99 | c1oi7A_ |
|
not modelled |
47.8 |
19 |
PDB header:synthetase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: the crystal structure of succinyl-coa synthetase alpha2 subunit from thermus thermophilus
|
|
|
| 100 | c2rqmA_ |
|
not modelled |
47.3 |
17 |
PDB header:chaperone
Chain: A: PDB Molecule:mesoderm development candidate 2;
PDBTitle: nmr solution structure of mesoderm development (mesd) - open2 conformation
|
|
|
| 101 | c2wtaA_ |
|
not modelled |
46.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
|
|
|
| 102 | c3tb4A_ |
|
not modelled |
46.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:vibriobactin-specific isochorismatase;
PDBTitle: crystal structure of the isc domain of vibb
|
|
|
| 103 | c3m4xA_ |
|
not modelled |
45.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:nol1/nop2/sun family protein;
PDBTitle: structure of a ribosomal methyltransferase
|
|
|
| 104 | c5zn8B_ |
|
not modelled |
45.1 |
21 |
PDB header:metal binding protein
Chain: B: PDB Molecule:isochorismatase;
PDBTitle: crystal structure of nicotinamidase pnca from bacillus subtilis
|
|
|
| 105 | c1yd2A_ |
|
not modelled |
44.9 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uvrabc system protein c;
PDBTitle: crystal structure of the giy-yig n-terminal endonuclease domain of2 uvrc from thermotoga maritima: point mutant y19f bound to the3 catalytic divalent cation
|
|
|
| 106 | c2csuB_ |
|
not modelled |
44.6 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
|
|
| 107 | d1aoga2 |
|
not modelled |
44.2 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 108 | d3c7bb2 |
|
not modelled |
44.2 |
25 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
|
|
| 109 | c2hxtA_ |
|
not modelled |
43.8 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:l-fuconate dehydratase;
PDBTitle: crystal structure of l-fuconate dehydratase from xanthomonas2 campestris liganded with mg++ and d-erythronohydroxamate
|
|
|
| 110 | d1q32a1 |
|
not modelled |
41.9 |
12 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Tyrosyl-DNA phosphodiesterase TDP1 |
|
|
| 111 | c2v4jA_ |
|
not modelled |
41.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
|
|
| 112 | d1nu5a1 |
|
not modelled |
40.9 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 113 | c1nu5A_ |
|
not modelled |
40.9 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:chloromuconate cycloisomerase;
PDBTitle: crystal structure of pseudomonas sp. p51 chloromuconate lactonizing2 enzyme
|
|
|
| 114 | c4zhjA_ |
|
not modelled |
40.4 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:mg-chelatase subunit chlh;
PDBTitle: crystal structure of the catalytic subunit of magnesium chelatase
|
|
|
| 115 | c2jlaD_ |
|
not modelled |
39.8 |
43 |
PDB header:transferase
Chain: D: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: crystal structure of e.coli mend, 2-succinyl-5-enolpyruvyl-2 6-hydroxy-3-cyclohexadiene-1-carboxylate synthase - semet3 protein
|
|
|
| 116 | c1pjtB_ |
|
not modelled |
39.7 |
18 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
| 117 | c1yd6A_ |
|
not modelled |
39.7 |
23 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uvrc;
PDBTitle: crystal structure of the giy-yig n-terminal endonuclease2 domain of uvrc from bacillus caldotenax
|
|
|
| 118 | c2zzxD_ |
|
not modelled |
39.5 |
8 |
PDB header:transport protein
Chain: D: PDB Molecule:abc transporter, solute-binding protein;
PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
|
|
|
| 119 | c3o93A_ |
|
not modelled |
39.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
|
|
|
| 120 | d1lvla2 |
|
not modelled |
38.3 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|