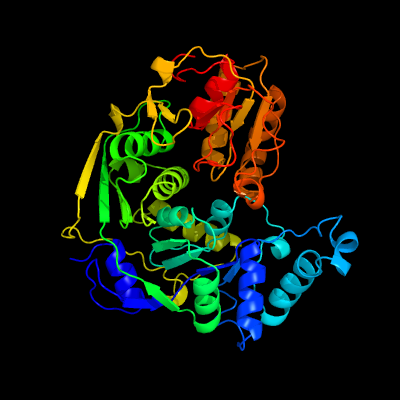
1 c4kp1A_
100.0
36
PDB header: isomeraseChain: A: PDB Molecule: isopropylmalate/citramalate isomerase large subunit;PDBTitle: crystal structure of ipm isomerase large subunit from methanococcus2 jannaschii (mj0499)
2 c4kp2A_
100.0
34
PDB header: lyaseChain: A: PDB Molecule: homoaconitase large subunit;PDBTitle: crystal structure of homoaconitase large subunit from methanococcus2 jannaschii (mj1003)
3 d1acoa2
100.0
26
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain
4 d1l5ja3
100.0
24
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain
5 c5acnA_
100.0
26
PDB header: lyase(carbon-oxygen)Chain: A: PDB Molecule: aconitase;PDBTitle: structure of activated aconitase. formation of the (4fe-4s) cluster in2 the crystal
6 d2b3ya2
100.0
22
Fold: Aconitase iron-sulfur domainSuperfamily: Aconitase iron-sulfur domainFamily: Aconitase iron-sulfur domain
7 c2b3yB_
100.0
22
PDB header: lyaseChain: B: PDB Molecule: iron-responsive element binding protein 1;PDBTitle: structure of a monoclinic crystal form of human cytosolic aconitase2 (irp1)
8 c1l5jB_
100.0
25
PDB header: lyaseChain: B: PDB Molecule: aconitate hydratase 2;PDBTitle: crystal structure of e. coli aconitase b.
9 c3bolB_
63.7
23
PDB header: transferaseChain: B: PDB Molecule: 5-methyltetrahydrofolate s-homocysteinePDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
10 d3bofa2
57.7
23
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase
11 c4cczA_
57.2
26
PDB header: transferaseChain: A: PDB Molecule: methionine synthase;PDBTitle: crystal structure of human 5-methyltetrahydrofolate-homocysteine2 methyltransferase, the homocysteine and folate binding domains
12 c4oc8A_
41.0
15
PDB header: hydrolaseChain: A: PDB Molecule: restriction endonuclease aspbhi;PDBTitle: dna modification-dependent restriction endonuclease aspbhi
13 c2e21A_
39.8
17
PDB header: ligaseChain: A: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils in a complex with amppnp from aquifex2 aeolicus.
14 c1ni5A_
37.8
15
PDB header: cell cycleChain: A: PDB Molecule: putative cell cycle protein mesj;PDBTitle: structure of the mesj pp-atpase from escherichia coli
15 c4yajA_
32.6
19
PDB header: ligaseChain: A: PDB Molecule: alpha subunit of acetyl-coenzyme a synthetasePDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
16 d1lt7a_
29.3
25
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase
17 c4rzlA_
27.5
7
PDB header: dna binding proteinChain: A: PDB Molecule: restriction endonuclease lpnpi;PDBTitle: dna recognition domain of the cytosine modification-dependent2 restriction endonuclease lpnpi
18 d1umya_
26.2
22
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase
19 c4oc8D_
25.3
17
PDB header: hydrolaseChain: D: PDB Molecule: restriction endonuclease aspbhi;PDBTitle: dna modification-dependent restriction endonuclease aspbhi
20 c5wusA_
24.5
31
PDB header: hydrolaseChain: A: PDB Molecule: chitinase;PDBTitle: crystal structure of a insect group iii chitinase (cad2) from ostrinia2 furnacalis
21 c3fseB_
not modelled
23.2
20
PDB header: hydrolaseChain: B: PDB Molecule: two-domain protein containing dj-1/thij/pfpi-like andPDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
22 c2gp4A_
not modelled
23.1
19
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
23 c5ze4A_
not modelled
20.7
15
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase, chloroplastic;PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
24 c2oxlA_
not modelled
20.7
47
PDB header: gene regulationChain: A: PDB Molecule: hypothetical protein ymgb;PDBTitle: structure and function of the e. coli protein ymgb: a protein critical2 for biofilm formation and acid resistance
25 c3a2kB_
not modelled
20.3
24
PDB header: ligase/rnaChain: B: PDB Molecule: trna(ile)-lysidine synthase;PDBTitle: crystal structure of tils complexed with trna
26 c5u32A_
not modelled
19.2
15
PDB header: transferaseChain: A: PDB Molecule: trna ligase;PDBTitle: crystal structure of fungal rna kinase
27 d1c0aa2
not modelled
18.6
21
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain
28 c5fifD_
not modelled
18.5
15
PDB header: ligaseChain: D: PDB Molecule: carboxylase;PDBTitle: carboxyltransferase domain of a single-chain bacterial carboxylase
29 c4l6wA_
not modelled
18.5
19
PDB header: ligaseChain: A: PDB Molecule: probable propionyl-coa carboxylase beta chain 6;PDBTitle: carboxyltransferase subunit (accd6) of mycobacterium tuberculosis2 acetyl-coa carboxylase
30 c2xivA_
not modelled
18.4
17
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of mycobacterium2 tuberculosis
31 c4rcnA_
not modelled
18.1
11
PDB header: ligaseChain: A: PDB Molecule: long-chain acyl-coa carboxylase;PDBTitle: structure and function of a single-chain, multi-domain long-chain2 acyl-coa carboxylase
32 d1ejba_
not modelled
17.9
14
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase
33 c2jz2A_
not modelled
17.6
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ssl0352 protein;PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
34 d2gp4a2
not modelled
16.4
19
Fold: IlvD/EDD N-terminal domain-likeSuperfamily: IlvD/EDD N-terminal domain-likeFamily: lvD/EDD N-terminal domain-like
35 c2kqvA_
not modelled
15.8
39
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 3;PDBTitle: sars coronavirus-unique domain (sud): three-domain molecular2 architecture in solution and rna binding. i: structure of the sud-m3 domain of sud-mc
36 c3uk7B_
not modelled
15.8
26
PDB header: transferaseChain: B: PDB Molecule: class i glutamine amidotransferase-like domain-containingPDBTitle: crystal structure of arabidopsis thaliana dj-1d
37 d1ni5a1
not modelled
15.5
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: PP-loop ATPase
38 c5mgyG_
not modelled
15.5
11
PDB header: protein bindingChain: G: PDB Molecule: fad:protein fmn transferase;PDBTitle: crystal structure of pseudomonas stutzeri flavinyl transferase apbe,2 apo form
39 c2yb1A_
not modelled
15.4
14
PDB header: hydrolaseChain: A: PDB Molecule: amidohydrolase;PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
40 d1oi4a1
not modelled
15.2
36
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI
41 c3iz5Q_
not modelled
14.8
9
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l5 (l18p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
42 c2gksB_
not modelled
13.0
17
PDB header: transferaseChain: B: PDB Molecule: bifunctional sat/aps kinase;PDBTitle: crystal structure of the bi-functional atp sulfurylase-aps kinase from2 aquifex aeolicus, a chemolithotrophic thermophile
43 c2jzfA_
not modelled
13.0
39
PDB header: viral proteinChain: A: PDB Molecule: replicase polyprotein 1ab;PDBTitle: nmr conformer closest to the mean coordinates of the domain 513-651 of2 the sars-cov nonstructural protein nsp3
44 c5uidC_
not modelled
12.5
13
PDB header: transferaseChain: C: PDB Molecule: aminotransferase tlmj;PDBTitle: the crystal structure of an aminotransferase tlmj from2 streptoalloteichus hindustanus
45 c4lkrA_
not modelled
12.4
16
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase deod-type;PDBTitle: crystal structure of deod-3 gene product from shewanella oneidensis2 mr-1, nysgrc target 029437
46 c1htrP_
not modelled
12.3
25
PDB header: aspartyl proteaseChain: P: PDB Molecule: progastricsin (pro segment);PDBTitle: crystal and molecular structures of human progastricsin at 1.622 angstroms resolution
47 c2x24B_
not modelled
12.1
16
PDB header: ligaseChain: B: PDB Molecule: acetyl-coa carboxylase;PDBTitle: bovine acc2 ct domain in complex with inhibitor
48 d1c41a_
not modelled
11.7
16
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase
49 c5yudA_
not modelled
11.6
26
PDB header: immune systemChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1e;PDBTitle: flagellin derivative in complex with the nlr protein naip5
50 c6b4cH_
not modelled
11.6
9
PDB header: antiviral proteinChain: H: PDB Molecule: viperin;PDBTitle: structure of viperin from trichoderma virens
51 c1x0uB_
not modelled
11.5
20
PDB header: lyaseChain: B: PDB Molecule: hypothetical methylmalonyl-coa decarboxylase alpha subunit;PDBTitle: crystal structure of the carboxyl transferase subunit of putative pcc2 of sulfolobus tokodaii
52 c4m70E_
not modelled
11.5
31
PDB header: plant proteinChain: E: PDB Molecule: ran gtpase activating protein 2;PDBTitle: crystal structure of potato rx-cc domain in complex with rangap2-wpp2 domain
53 c4mnpA_
not modelled
11.4
11
PDB header: sugar binding proteinChain: A: PDB Molecule: n-acetylneuraminate-binding protein;PDBTitle: structure of the sialic acid binding protein from fusobacterium2 nucleatum subsp. nucleatum atcc 25586
54 c5jw6A_
not modelled
11.3
19
PDB header: oxidoreductaseChain: A: PDB Molecule: aspartate-semialdehyde dehydrogenase;PDBTitle: cystal structure of aspartate semialdehyde dehydrogenase from2 aspergillus fumigatus
55 c2w9mB_
not modelled
10.9
20
PDB header: dna replicationChain: B: PDB Molecule: polymerase x;PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
56 c3h8aF_
not modelled
10.9
27
PDB header: lyase/protein bindingChain: F: PDB Molecule: rnase e;PDBTitle: crystal structure of e. coli enolase bound to its cognate rnase e2 recognition domain
57 c1xovA_
not modelled
10.8
16
PDB header: hydrolaseChain: A: PDB Molecule: ply protein;PDBTitle: the crystal structure of the listeria monocytogenes bacteriophage psa2 endolysin plypsa
58 c3qpbB_
not modelled
10.8
18
PDB header: transferaseChain: B: PDB Molecule: uridine phosphorylase;PDBTitle: crystal structure of streptococcus pyogenes uridine phosphorylase2 reveals a subclass of the np-i superfamily
59 c2o2qA_
not modelled
10.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: formyltetrahydrofolate dehydrogenase;PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp
60 c5kf6B_
not modelled
10.6
17
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of proline utilization a from sinorhizobium meliloti2 complexed with l-tetrahydrofuroic acid and nad+ in space group p21
61 c3h8aE_
not modelled
10.3
27
PDB header: lyase/protein bindingChain: E: PDB Molecule: rnase e;PDBTitle: crystal structure of e. coli enolase bound to its cognate rnase e2 recognition domain
62 c3io1B_
not modelled
10.3
17
PDB header: hydrolaseChain: B: PDB Molecule: aminobenzoyl-glutamate utilization protein;PDBTitle: crystal structure of aminobenzoyl-glutamate utilization2 protein from klebsiella pneumoniae
63 c4binA_
not modelled
10.3
12
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase amic;PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
64 d1sqda2
not modelled
10.1
28
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases
65 c6b5bA_
not modelled
10.1
26
PDB header: immune systemChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1e;PDBTitle: cryo-em structure of the naip5-nlrc4-flagellin inflammasome
66 d1ppya_
not modelled
10.1
19
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC
67 d2a7sa1
not modelled
10.1
15
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain
68 d2ac7a1
not modelled
10.0
15
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases
69 c3pbiA_
not modelled
9.9
19
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
70 c5d3aA_
not modelled
9.8
50
PDB header: motor proteinChain: A: PDB Molecule: kinesin-like protein kif21a;PDBTitle: kif21a regulatory coiled coil
71 c3v9iD_
not modelled
9.7
9
PDB header: oxidoreductaseChain: D: PDB Molecule: delta-1-pyrroline-5-carboxylate dehydrogenase,PDBTitle: crystal structure of human 1-pyrroline-5-carboxylate dehydrogenase2 mutant s352l
72 c2nvgA_
not modelled
9.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: soluble domain of rieske iron sulfur protein.
73 d1v8ba2
not modelled
9.6
37
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase
74 c3hskB_
not modelled
9.5
13
PDB header: oxidoreductaseChain: B: PDB Molecule: aspartate-semialdehyde dehydrogenase;PDBTitle: crystal structure of aspartate semialdehyde dehydrogenase with nadp2 from candida albicans
75 c4xb6D_
not modelled
9.5
14
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
76 c5ip4E_
not modelled
9.4
16
PDB header: transferaseChain: E: PDB Molecule: neuronal migration protein doublecortin;PDBTitle: x-ray structure of the c-terminal domain of human doublecortin
77 c5ux5C_
not modelled
9.4
23
PDB header: oxidoreductase/transferaseChain: C: PDB Molecule: bifunctional protein proline utilization a (puta);PDBTitle: structure of proline utilization a (puta) from corynebacterium2 freiburgense
78 c2ebbA_
not modelled
9.3
15
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
79 c2fgxA_
not modelled
9.2
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thioredoxin;PDBTitle: solution nmr structure of protein ne2328 from nitrosomonas2 europaea. northeast structural genomics consortium target3 net3.
80 c4pfrB_
not modelled
9.1
18
PDB header: solute-binding proteinChain: B: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 rhodobacter sphaeroides (rsph17029_3541, target efi-510203), apo open3 partially disordered
81 c6hlwB_
not modelled
9.1
29
PDB header: viral proteinChain: B: PDB Molecule: genome polyprotein;PDBTitle: crystal structure of human acbd3 gold domain in complex with 3a2 protein of enterovirus-a71 (fusion protein)
82 d1obra_
not modelled
9.0
33
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Carboxypeptidase T
83 c1od4C_
not modelled
9.0
16
PDB header: ligaseChain: C: PDB Molecule: acetyl-coenzyme a carboxylase;PDBTitle: acetyl-coa carboxylase carboxyltransferase domain
84 c3gyyC_
not modelled
9.0
27
PDB header: transport proteinChain: C: PDB Molecule: periplasmic substrate binding protein;PDBTitle: the ectoine binding protein of the teaabc trap transporter teaa in the2 apo-state
85 c4m3nA_
not modelled
8.9
12
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase deod-type;PDBTitle: crystal structure of purine nucleoside phosphorylase from meiothermus2 ruber dsm 1279, nysgrc target 029804.
86 c5ingC_
not modelled
8.8
12
PDB header: transferaseChain: C: PDB Molecule: putative carboxyl transferase;PDBTitle: a crotonyl-coa reductase-carboxylase independent pathway for assembly2 of unusual alkylmalonyl-coa polyketide synthase extender unit
87 d2ga5a1
not modelled
8.7
29
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
88 c2k89A_
not modelled
8.7
21
PDB header: protein bindingChain: A: PDB Molecule: phospholipase a-2-activating protein;PDBTitle: solution structure of a novel ubiquitin-binding domain from2 human plaa (pfuc, gly76-pro77 cis isomer)
89 d1f0xa1
not modelled
8.7
11
Fold: Ferredoxin-likeSuperfamily: FAD-linked oxidases, C-terminal domainFamily: D-lactate dehydrogenase
90 d1wvfa1
not modelled
8.6
11
Fold: Ferredoxin-likeSuperfamily: FAD-linked oxidases, C-terminal domainFamily: Vanillyl-alcohol oxidase-like
91 d1woha_
not modelled
8.6
28
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Arginase-like amidino hydrolases
92 d1sp8a2
not modelled
8.5
15
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases
93 c4jc0B_
not modelled
8.5
14
PDB header: transferaseChain: B: PDB Molecule: ribosomal protein s12 methylthiotransferase rimo;PDBTitle: crystal structure of thermotoga maritima holo rimo in complex with2 pentasulfide, northeast structural genomics consortium target vr77
94 d2byea1
not modelled
8.5
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD
95 c1g8gB_
not modelled
8.3
13
PDB header: transferaseChain: B: PDB Molecule: sulfate adenylyltransferase;PDBTitle: atp sulfurylase from s. cerevisiae: the binary product complex with2 aps
96 d1iuka_
not modelled
8.3
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
97 c6hmvB_
not modelled
8.3
21
PDB header: viral proteinChain: B: PDB Molecule: genome polyprotein;PDBTitle: crystal structure of human acbd3 gold domain in complex with 3a2 protein of enterovirus-d68 (fusion protein, lvvy mutant)
98 c4uoiB_
not modelled
8.2
28
PDB header: viral proteinChain: B: PDB Molecule: genome polyprotein;PDBTitle: unexpected structure for the n-terminal domain of hepatitis c virus2 envelope glycoprotein e1
99 d1skye2
not modelled
8.1
17
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase