| 1 | d1w0da_
|
|
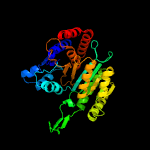 |
100.0 |
100 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 2 | c2d1cB_
|
|
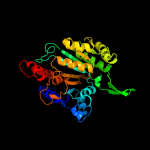 |
100.0 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
|
|
|
| 3 | d1pb1a_
|
|
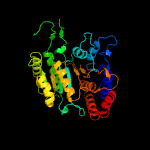 |
100.0 |
32 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 4 | c3fmxX_
|
|
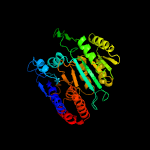 |
100.0 |
41 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas putida2 complexed with nadh
|
|
|
|
| 5 | c2d4vD_
|
|
 |
100.0 |
32 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
|
|
|
| 6 | d1cm7a_
|
|
 |
100.0 |
41 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 7 | d1v53a1
|
|
 |
100.0 |
39 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 8 | d1g2ua_
|
|
 |
100.0 |
43 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 9 | d1cnza_
|
|
 |
100.0 |
41 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 10 | d1a05a_
|
|
 |
100.0 |
42 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 11 | d1hqsa_
|
|
 |
100.0 |
34 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 12 | d1xaca_
|
|
 |
100.0 |
42 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 13 | c4iwhA_
|
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of a 3-isopropylmalate dehydrogenase from2 burkholderia pseudomallei
|
|
|
|
| 14 | d1vlca_
|
|
 |
100.0 |
41 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 15 | c3r8wC_
|
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
|
|
|
| 16 | c2e0cA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
|
|
|
| 17 | c3uduG_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of putative 3-isopropylmalate dehydrogenase from2 campylobacter jejuni
|
|
|
|
| 18 | c3vl3A_
|
|
 |
100.0 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: 3-isopropylmalate dehydrogenase from shewanella oneidensis mr-1 at 3402 mpa
|
|
|
|
| 19 | c3ty3A_
|
|
 |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable homoisocitrate dehydrogenase;
PDBTitle: crystal structure of homoisocitrate dehydrogenase from2 schizosaccharomyces pombe bound to glycyl-glycyl-glycine
|
|
|
|
| 20 | c3u1hA_
|
|
 |
100.0 |
41 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
|
|
|
| 21 | d1wpwa_ |
|
not modelled |
100.0 |
35 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 22 | c1x0lB_ |
|
not modelled |
100.0 |
43 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
|
|
| 23 | c5grhA_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nad] subunit alpha,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
| 24 | c3blxL_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
| 25 | c5grhB_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase [nad] subunit gamma,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
| 26 | c5hn6A_ |
|
not modelled |
100.0 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of beta-decarboxylating dehydrogenase (tk0280) from2 thermococcus kodakarensis complexed with mn and 3-isopropylmalate
|
|
|
| 27 | c3blxM_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
| 28 | c1zorB_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
|
|
| 29 | c1tyoA_ |
|
not modelled |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
|
|
| 30 | c2qfyE_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
|
|
| 31 | c2uxqB_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
|
|
| 32 | d1lwda_ |
|
not modelled |
100.0 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 33 | d1t0la_ |
|
not modelled |
100.0 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 34 | c3us8A_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
|
|
| 35 | c2iv0A_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
|
|
| 36 | c4aoyD_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: open ctidh. the complex structures of isocitrate dehydrogenase from2 clostridium thermocellum and desulfotalea psychrophila, support a new3 active site locking mechanism
|
|
|
| 37 | d1r8ka_ |
|
not modelled |
97.6 |
27 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 38 | c2b0tA_ |
|
not modelled |
97.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp isocitrate dehydrogenase;
PDBTitle: structure of monomeric nadp isocitrate dehydrogenase
|
|
|
| 39 | c4jqpA_ |
|
not modelled |
97.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase;
PDBTitle: x-ray crystal structure of a 4-hydroxythreonine-4-phosphate2 dehydrogenase from burkholderia phymatum
|
|
|
| 40 | c6g3uA_ |
|
not modelled |
96.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: structure of pseudomonas aeruginosa isocitrate dehydrogenase, idh
|
|
|
| 41 | d1itwa_ |
|
not modelled |
96.8 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Monomeric isocitrate dehydrogenase |
|
|
| 42 | c2hi1A_ |
|
not modelled |
96.7 |
34 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
|
|
| 43 | c4atyA_ |
|
not modelled |
96.6 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:terephthalate 1,2-cis-dihydrodiol dehydrogenase;
PDBTitle: crystal structure of a terephthalate 1,2-cis-2 dihydrodioldehydrogenase from burkholderia xenovorans3 lb400
|
|
|
| 44 | d1ptma_ |
|
not modelled |
96.4 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 45 | c1yxoB_ |
|
not modelled |
96.4 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 1;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
|
|
|
| 46 | c3tsnD_ |
|
not modelled |
96.1 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase;
PDBTitle: 4-hydroxythreonine-4-phosphate dehydrogenase from campylobacter jejuni
|
|
|
| 47 | c3s40C_ |
|
not modelled |
60.6 |
9 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
|
|
| 48 | c6d6yA_ |
|
not modelled |
56.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:apra methyltransferase 2;
PDBTitle: apra methyltransferase 2 - gnat didomain in complex with sah
|
|
|
| 49 | c3cfxA_ |
|
not modelled |
53.0 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein ma_0280;
PDBTitle: crystal structure of m. acetivorans periplasmic binding protein2 moda/wtpa with bound tungstate
|
|
|
| 50 | d3thia_ |
|
not modelled |
52.5 |
22 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 51 | c3k6wA_ |
|
not modelled |
47.2 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:solute-binding protein ma_0280;
PDBTitle: apo and ligand bound structures of moda from the archaeon2 methanosarcina acetivorans
|
|
|
| 52 | d2fpoa1 |
|
not modelled |
44.6 |
11 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
| 53 | d2onsa1 |
|
not modelled |
44.6 |
21 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 54 | c1orhA_ |
|
not modelled |
42.1 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:protein arginine n-methyltransferase 1;
PDBTitle: structure of the predominant protein arginine methyltransferase prmt1
|
|
|
| 55 | c4c0xA_ |
|
not modelled |
41.9 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fmn-dependent nadh-azoreductase 1;
PDBTitle: the crystal strucuture of ppazor in complex with anthraquinone-2-2 sulfonate
|
|
|
| 56 | c5f4bB_ |
|
not modelled |
41.9 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad(p)h dehydrogenase (quinone);
PDBTitle: structure of b. abortus wrba-related protein a (wrpa)
|
|
|
| 57 | c5odhH_ |
|
not modelled |
39.6 |
22 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:heterodisulfide reductase, subunit b;
PDBTitle: heterodisulfide reductase / [nife]-hydrogenase complex from2 methanothermococcus thermolithotrophicus soaked with heterodisulfide3 for 3.5 minutes
|
|
|
| 58 | c4kysA_ |
|
not modelled |
39.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine pyridinylase i;
PDBTitle: clostridium botulinum thiaminase i in complex with thiamin
|
|
|
| 59 | d1oria_ |
|
not modelled |
36.2 |
5 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
|
|
| 60 | c2fyuE_ |
|
not modelled |
35.0 |
26 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
|
|
| 61 | c1p84E_ |
|
not modelled |
34.8 |
32 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
|
|
| 62 | c5xtuA_ |
|
not modelled |
34.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdsl-family esterase;
PDBTitle: crystal structure of gdsl esterase of photobacterium sp. j15
|
|
|
| 63 | c6ccaA_ |
|
not modelled |
32.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:disa protein;
PDBTitle: crystal structure of dsza carbon methyltransferase
|
|
|
| 64 | d2fiqa1 |
|
not modelled |
32.5 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
|
|
| 65 | d1riea_ |
|
not modelled |
32.1 |
26 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 66 | c4p7cB_ |
|
not modelled |
31.5 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:trna (mo5u34)-methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from pseudomonas2 syringae pv. tomato
|
|
|
| 67 | c3cfzA_ |
|
not modelled |
31.0 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:upf0100 protein mj1186;
PDBTitle: crystal structure of m. jannaschii periplasmic binding2 protein moda/wtpa with bound tungstate
|
|
|
| 68 | c5mkbF_ |
|
not modelled |
30.4 |
25 |
PDB header:sugar binding protein
Chain: F: PDB Molecule:male1;
PDBTitle: maltodextrin binding protein male1 from l. casei bl23 without ligand
|
|
|
| 69 | c4ptzC_ |
|
not modelled |
30.0 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:fmn reductase ssue;
PDBTitle: crystal structure of the escherichia coli alkanesulfonate fmn2 reductase ssue in fmn-bound form
|
|
|
| 70 | c4ycsC_ |
|
not modelled |
29.6 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative lipoprotein from peptoclostridium2 difficile 630 (fragment)
|
|
|
| 71 | c5ygeA_ |
|
not modelled |
29.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:amino-acid acetyltransferase;
PDBTitle: arga complexed with acecoa and glutamate
|
|
|
| 72 | c1twyG_ |
|
not modelled |
26.2 |
18 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
|
|
|
| 73 | d1twya_ |
|
not modelled |
26.2 |
18 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 74 | d1ydga_ |
|
not modelled |
26.2 |
24 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
|
|
| 75 | c6fuvA_ |
|
not modelled |
25.9 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:solute binding protein, blmnbp1 in complex with
PDBTitle: structure of a manno-oligosaccharide specific solute binding protein,2 blmnbp2 from bifidobacterium animalis subsp. lactis atcc 27673 in3 complex with mannotriose
|
|
|
| 76 | c5f7vA_ |
|
not modelled |
25.6 |
21 |
PDB header:cycloalternan binding protein
Chain: A: PDB Molecule:lmo0181 protein;
PDBTitle: abc substrate-binding protein lmo0181 from listeria monocytogenes in2 complex with cycloalternan
|
|
|
| 77 | d2esra1 |
|
not modelled |
25.2 |
16 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
| 78 | c2k4mA_ |
|
not modelled |
24.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein mth_1000,2 northeast structural genomics consortium target tr8
|
|
|
| 79 | c4r81C_ |
|
not modelled |
24.4 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase;
PDBTitle: nad(p)h:quinone oxidoreductase from methanothermobacter marburgensis
|
|
|
| 80 | c4gd5B_ |
|
not modelled |
23.9 |
9 |
PDB header:transport protein
Chain: B: PDB Molecule:phosphate abc transporter, phosphate-binding protein;
PDBTitle: x-ray crystal structure of a putative phosphate abc transporter2 substrate-binding protein with bound phosphate from clostridium3 perfringens
|
|
|
| 81 | c5lkjA_ |
|
not modelled |
23.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:histone-arginine methyltransferase carm1;
PDBTitle: crystal structure of mouse carm1 in complex with ligand sa684
|
|
|
| 82 | c2pebB_ |
|
not modelled |
23.4 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative dioxygenase;
PDBTitle: crystal structure of a putative dioxygenase (npun_f1925) from nostoc2 punctiforme pcc 73102 at 1.46 a resolution
|
|
|
| 83 | d3cx5e1 |
|
not modelled |
23.1 |
32 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 84 | c3pvsA_ |
|
not modelled |
23.0 |
25 |
PDB header:recombination
Chain: A: PDB Molecule:replication-associated recombination protein a;
PDBTitle: structure and biochemical activities of escherichia coli mgsa
|
|
|
| 85 | c3lcmB_ |
|
not modelled |
21.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
|
|
| 86 | c6fjlD_ |
|
not modelled |
21.8 |
13 |
PDB header:metal binding protein
Chain: D: PDB Molecule:abc-type fe3+ transport system, periplasmic component;
PDBTitle: structure of ibps from dickeya dadantii
|
|
|
| 87 | d2r9ga1 |
|
not modelled |
21.6 |
31 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:MgsA/YrvN C-terminal domain-like |
|
|
| 88 | d1j08a1 |
|
not modelled |
20.4 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:PDI-like |
|
|
| 89 | d2fyta1 |
|
not modelled |
20.1 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
|
|
| 90 | c4r6hA_ |
|
not modelled |
19.6 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:solute binding protein msme;
PDBTitle: crystal structure of putative binding protein msme from bacillus2 subtilis subsp. subtilis str. 168, target efi-510764, an open3 conformation
|
|
|
| 91 | d1qapa1 |
|
not modelled |
19.4 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
| 92 | c6dk9I_ |
|
not modelled |
18.8 |
23 |
PDB header:lyase
Chain: I: PDB Molecule:dna damage-inducible protein;
PDBTitle: yeast ddi2 cyanamide hydratase
|
|
|
| 93 | d3bgea1 |
|
not modelled |
18.4 |
22 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:MgsA/YrvN C-terminal domain-like |
|
|
| 94 | c5ftwA_ |
|
not modelled |
18.0 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:chemotaxis protein methyltransferase;
PDBTitle: crystal structure of glutamate o-methyltransferase in2 complex with s- adenosyl-l-homocysteine (sah) from3 bacillus subtilis
|
|
|
| 95 | c5thyB_ |
|
not modelled |
17.9 |
19 |
PDB header:transferase,lyase
Chain: B: PDB Molecule:curj;
PDBTitle: crystal structure of semet-substituted curj carbon methyltransferase
|
|
|
| 96 | c3b3jA_ |
|
not modelled |
17.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:histone-arginine methyltransferase carm1;
PDBTitle: the 2.55 a crystal structure of the apo catalytic domain of2 coactivator-associated arginine methyl transferase i(carm1:28-507,3 residues 28-146 and 479-507 not ordered)
|
|
|
| 97 | c2qhxB_ |
|
not modelled |
17.6 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pteridine reductase 1;
PDBTitle: structure of pteridine reductase from leishmania major2 complexed with a ligand
|
|
|
| 98 | c4hcwC_ |
|
not modelled |
17.2 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:thiaminase-i;
PDBTitle: structure of a eukaryotic thiaminase-i
|
|
|
| 99 | d1g6q1_ |
|
not modelled |
17.1 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Arginine methyltransferase |
|
|







































































































































































































































