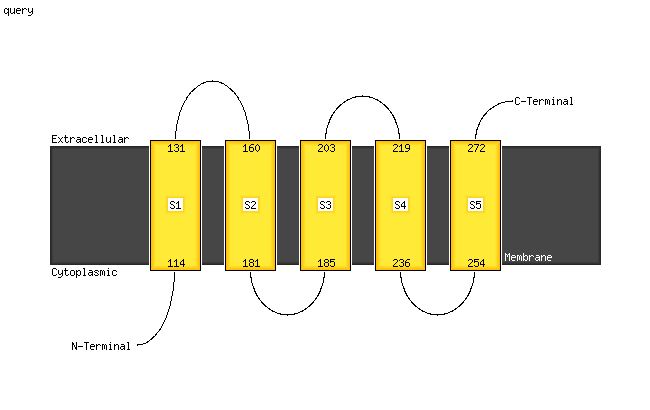
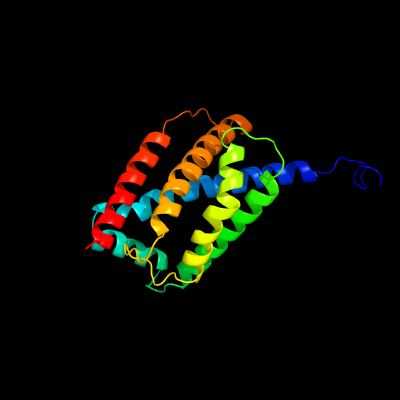
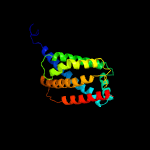
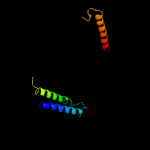
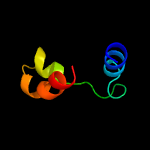
1 d2bs2c1
55.0
13
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Fumarate reductase respiratory complex cytochrome b subunit, FrdC
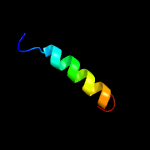
2 c5gjqZ_
47.9
16
PDB header: hydrolaseChain: Z: PDB Molecule: 26s proteasome non-atpase regulatory subunit 2;PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
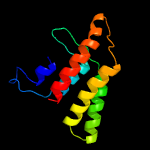
3 c6hwhB_
34.4
15
PDB header: electron transportChain: B: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
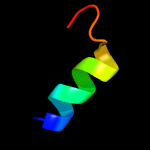
4 c5khrA_
34.3
33
PDB header: cell cycleChain: A: PDB Molecule: anaphase-promoting complex subunit 1;PDBTitle: model of human anaphase-promoting complex/cyclosome complex (apc152 deletion mutant) in complex with the e2 ube2c/ubch10 poised for3 ubiquitin ligation to substrate (apc/c-cdc20-substrate-ube2c)
5 c5a31A_
28.3
37
PDB header: cell cycleChain: A: PDB Molecule: anaphase-promoting complex subunit 1;PDBTitle: structure of the human apc-cdh1-hsl1-ubch10 complex.
6 c4ui9A_
27.5
37
PDB header: cell cycleChain: A: PDB Molecule: anaphase-promoting complex subunit 1;PDBTitle: atomic structure of the human anaphase-promoting complex
7 c2lzsE_
26.7
21
PDB header: protein transportChain: E: PDB Molecule: sec-independent protein translocase protein tata;PDBTitle: tata oligomer
8 c5xj6A_
26.4
19
PDB header: transferaseChain: A: PDB Molecule: glycerol-3-phosphate acyltransferase;PDBTitle: crystal structure of plsy (ygih), an integral membrane glycerol 3-2 phosphate acyltransferase - the glycerol 3-phosphate form
9 c2mi2A_
25.0
29
PDB header: transport proteinChain: A: PDB Molecule: sec-independent protein translocase protein tatb;PDBTitle: solution structure of the e. coli tatb protein in dpc micelles
10 c5khuA_
24.7
37
PDB header: cell cycleChain: A: PDB Molecule: anaphase-promoting complex subunit 1;PDBTitle: model of human anaphase-promoting complex/cyclosome (apc15 deletion2 mutant), in complex with the mitotic checkpoint complex (apc/c-cdc20-3 mcc) based on cryo em data at 4.8 angstrom resolution
11 c5awwG_
23.1
19
PDB header: protein transport/immune systemChain: G: PDB Molecule: putative preprotein translocase, secg subunit;PDBTitle: precise resting state of thermus thermophilus secyeg
12 c3vrbG_
21.2
18
PDB header: oxidoreductase/oxidoreductase inhibitorChain: G: PDB Molecule: cytochrome b-large subunit;PDBTitle: mitochondrial rhodoquinol-fumarate reductase from the parasitic2 nematode ascaris suum with the specific inhibitor flutolanil and3 substrate fumarate
13 c5l9uA_
20.9
35
PDB header: signaling proteinChain: A: PDB Molecule: anaphase-promoting complex subunit 1;PDBTitle: model of human anaphase-promoting complex/cyclosome (apc/c-cdh1) with2 a cross linked ubiquitin variant-substrate-ube2c (ubch10) complex3 representing key features of multiubiquitination
14 c4ezdB_
20.2
20
PDB header: transport proteinChain: B: PDB Molecule: urea transporter 1;PDBTitle: crystal structure of the ut-b urea transporter from bos taurus bound2 to selenourea
15 c6iu3A_
19.7
16
PDB header: metal transportChain: A: PDB Molecule: vit1;PDBTitle: crystal structure of iron transporter vit1 with zinc ions
16 c5xmjG_
19.2
13
PDB header: electron transportChain: G: PDB Molecule: fumarate reductase respiratory complex;PDBTitle: crystal structure of quinol:fumarate reductase from desulfovibrio2 gigas
17 c2lcnA_
19.1
31
PDB header: membrane proteinChain: A: PDB Molecule: walp19-p10 peptide;PDBTitle: 1h and 15n assignments of walp19-p10 peptide in sds micelles
18 d1jvra_
15.3
21
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: HTLV-II matrix protein
19 c2lcoA_
14.7
31
PDB header: membrane proteinChain: A: PDB Molecule: walp19-p8 peptide;PDBTitle: 1h and 15n assignments of walp19-p8 peptide in sds micelles
20 c2l16A_
12.7
17
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
21 c2k9yB_
not modelled
12.3
29
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
22 c2k9yA_
not modelled
11.5
29
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
23 c3k3gA_
not modelled
11.2
24
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
24 c5sxpF_
not modelled
7.7
47
PDB header: signaling protein/ligaseChain: F: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
25 c6ffvA_
not modelled
7.3
11
PDB header: transport proteinChain: A: PDB Molecule: btum;PDBTitle: the crystal structure of btum cobalamin transporter
26 c5l6nI_
not modelled
7.2
27
PDB header: hydrolaseChain: I: PDB Molecule: thrombin inhibitor madanin 1;PDBTitle: disulfated madanin-thrombin complex
27 c5ldwc_
not modelled
6.9
11
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class1
28 c5lc5c_
not modelled
6.9
11
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class2
29 c2miiA_
not modelled
6.9
12
PDB header: protein bindingChain: A: PDB Molecule: penicillin-binding protein activator lpob;PDBTitle: nmr structure of e. coli lpob
30 d3boea1
not modelled
6.9
33
Fold: CdCA1 repeat-likeSuperfamily: CdCA1 repeat-likeFamily: CdCA1 repeat-like
31 d1xm5a_
not modelled
6.3
36
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Predicted metal-dependent hydrolase
32 c3rkoK_
not modelled
6.2
23
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit k;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
33 c5sxpG_
not modelled
6.2
44
PDB header: signaling protein/ligaseChain: G: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
34 c2cpbA_
not modelled
6.0
11
PDB header: viral proteinChain: A: PDB Molecule: m13 major coat protein;PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
35 c3ipkA_
not modelled
5.8
7
PDB header: cell adhesionChain: A: PDB Molecule: agi/ii;PDBTitle: crystal structure of a3vp1 of agi/ii of streptococcus mutans
36 c1xaxA_
not modelled
5.8
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0054 protein hi0004;PDBTitle: nmr structure of hi0004, a putative essential gene product2 from haemophilus influenzae
37 c4heaK_
not modelled
5.5
18
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit 11;PDBTitle: crystal structure of the entire respiratory complex i from thermus2 thermophilus
38 c5azcA_
not modelled
5.4
16
PDB header: transferaseChain: A: PDB Molecule: prolipoprotein diacylglyceryl transferase;PDBTitle: crystal structure of escherichia coli lgt in complex with2 phosphatidylglycerol
39 c4gvsA_
not modelled
5.4
22
PDB header: hydrolaseChain: A: PDB Molecule: methenyltetrahydromethanopterin cyclohydrolase;PDBTitle: x-ray structure of the archaeoglobus fulgidus methenyl-2 tetrahydromethanopterin cyclohydrolase in complex with n5-formyl-3 tetrahydromethanopterin
40 c4cdiC_
not modelled
5.3
23
PDB header: membrane proteinChain: C: PDB Molecule: predicted protein;PDBTitle: crystal structure of acrb-acrz complex
41 c4cz8A_
not modelled
5.3
6
PDB header: membrane proteinChain: A: PDB Molecule: na+/h+ antiporter, putative;PDBTitle: structure of the sodium proton antiporter panhap from2 pyrococcus abyssii at ph 8.
42 c5z62N_
not modelled
5.3
29
PDB header: electron transportChain: N: PDB Molecule: cytochrome c oxidase subunit ndufa4;PDBTitle: structure of human cytochrome c oxidase
43 c6cfwI_
not modelled
5.1
27
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase