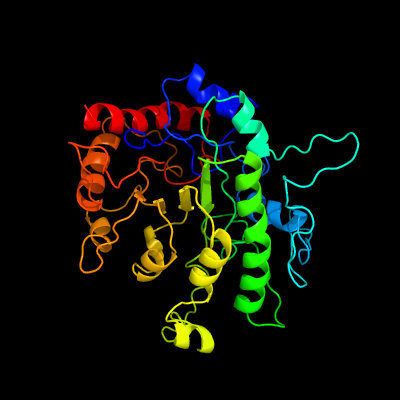
| 1 | c1u22A_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--
PDBTitle: a. thaliana cobalamine independent methionine synthase
|
|
|
|
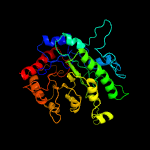
| 2 | c1t7lA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of cobalamin-independent methionine synthase from t.2 maritima
|
|
|
|
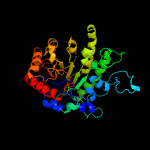
| 3 | c4ztxA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:cobalamin-independent methionine synthase;
PDBTitle: neurospora crassa cobalamin-independent methionine synthase complexed2 with zn2+
|
|
|
|
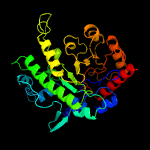
| 4 | c3l7sA_
|
|
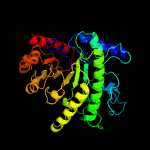 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of mete coordinated with zinc from streptococcus2 mutans
|
|
|
|
| 5 | c2nq5A_
|
|
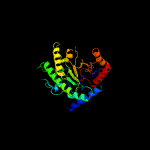 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of methyltransferase from streptococcus mutans
|
|
|
|
| 6 | d1u1ha2
|
|
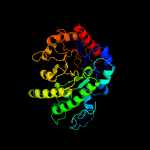 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
|
|
|
| 7 | c3ppgA_
|
|
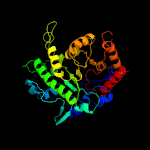 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydropteroyltriglutamate--homocysteine
PDBTitle: crystal structure of the candida albicans methionine synthase by2 surface entropy reduction, alanine variant with zinc
|
|
|
|
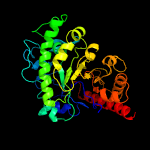
| 8 | c3rpdB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:methionine synthase (b12-independent);
PDBTitle: the structure of a b12-independent methionine synthase from shewanella2 sp. w3-18-1 in complex with selenomethionine.
|
|
|
|
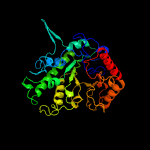
| 9 | c1ypxA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative vitamin-b12 independent methionine synthase family
PDBTitle: crystal structure of the putative vitamin-b12 independent methionine2 synthase from listeria monocytogenes, northeast structural genomics3 target lmr13
|
|
|
|
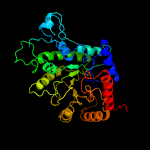
| 10 | d1u1ha1
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Cobalamin-independent methionine synthase |
|
|
|
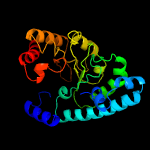
| 11 | c4ay8B_
|
|
 |
99.7 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:methylcobalamin\: coenzyme m methyltransferase;
PDBTitle: semet-derivative of a methyltransferase from m. mazei
|
|
|
|
| 12 | d1j93a_
|
|
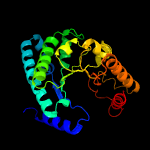 |
99.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
|
| 13 | c3cyvA_
|
|
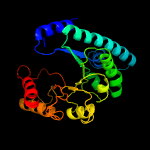 |
99.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
|
|
|
| 14 | c4exqA_
|
|
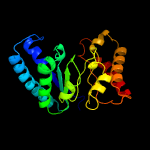 |
99.6 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase (upd) from2 burkholderia thailandensis e264
|
|
|
|
| 15 | c1jpkA_
|
|
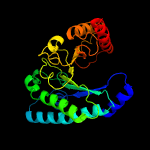 |
99.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
|
|
|
| 16 | c4zr8B_
|
|
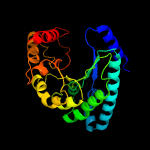 |
99.5 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: structure of uroporphyrinogen decarboxylase from acinetobacter2 baumannii
|
|
|
|
| 17 | d1r3sa_
|
|
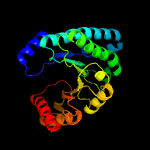 |
99.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
|
| 18 | c2infB_
|
|
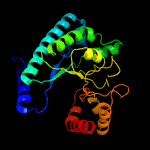 |
99.5 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from bacillus2 subtilis
|
|
|
|
| 19 | c2ejaB_
|
|
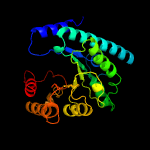 |
99.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
|
|
|
| 20 | c3rhgA_
|
|
 |
86.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phophotriesterase;
PDBTitle: crystal structure of amidohydrolase pmi1525 (target efi-500319) from2 proteus mirabilis hi4320
|
|
|
|
| 21 | d1ajza_ |
|
not modelled |
83.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 22 | c5odcD_ |
|
not modelled |
83.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:methyl-viologen reducing hydrogenase subunit d;
PDBTitle: heterodisulfide reductase / [nife]-hydrogenase complex from2 methanothermococcus thermolithotrophicus at 2.3 a resolution
|
|
|
| 23 | d1rvga_ |
|
not modelled |
81.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 24 | d1gvfa_ |
|
not modelled |
79.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 25 | c3qc3B_ |
|
not modelled |
74.9 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
|
|
| 26 | d1h1ya_ |
|
not modelled |
74.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 27 | c6omzA_ |
|
not modelled |
74.7 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from mycobacterium2 smegmatis with bound 6-hydroxymethylpterin-monophosphate
|
|
|
| 28 | d1tqxa_ |
|
not modelled |
74.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 29 | c1tx2A_ |
|
not modelled |
73.1 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
|
|
| 30 | d1tx2a_ |
|
not modelled |
73.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 31 | c6ofuC_ |
|
not modelled |
72.3 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:ydji aldolase;
PDBTitle: x-ray crystal structure of the ydji aldolase from escherichia coli k12
|
|
|
| 32 | d1f6ya_ |
|
not modelled |
72.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
|
|
| 33 | d1rpxa_ |
|
not modelled |
71.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 34 | c3k2gA_ |
|
not modelled |
70.5 |
15 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
|
|
| 35 | c3ct7E_ |
|
not modelled |
70.3 |
16 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
|
|
| 36 | c3c52B_ |
|
not modelled |
65.3 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from helicobacter pylori2 in complex with phosphoglycolohydroxamic acid, a competitive3 inhibitor
|
|
|
| 37 | c5u4nA_ |
|
not modelled |
65.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-1;
PDBTitle: crystal structure of a fructose-bisphosphate aldolase from neisseria2 gonorrhoeae
|
|
|
| 38 | c2vp8A_ |
|
not modelled |
64.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
|
|
| 39 | d1tqja_ |
|
not modelled |
64.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 40 | c2bmbA_ |
|
not modelled |
58.2 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase from3 saccharomyces cerevisiae
|
|
|
| 41 | c3tr9A_ |
|
not modelled |
56.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
|
|
| 42 | c4mwaA_ |
|
not modelled |
56.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
| 43 | c2iswB_ |
|
not modelled |
55.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
|
|
| 44 | c4nu7C_ |
|
not modelled |
53.9 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 45 | c3q94B_ |
|
not modelled |
52.1 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
|
|
| 46 | c3inpA_ |
|
not modelled |
51.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
|
|
| 47 | d1xrta2 |
|
not modelled |
47.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Hydantoinase (dihydropyrimidinase), catalytic domain |
|
|
| 48 | c5umfB_ |
|
not modelled |
47.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: crystal structure of a ribulose-phosphate 3-epimerase from neisseria2 gonorrhoeae with bound phosphate
|
|
|
| 49 | d1ad1a_ |
|
not modelled |
46.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 50 | c5uurA_ |
|
not modelled |
45.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: xanthomonas albilineans dihydropteroate synthase with 4-aminobenzoic2 acid
|
|
|
| 51 | c2y5sA_ |
|
not modelled |
43.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
|
|
| 52 | c2yciX_ |
|
not modelled |
43.1 |
17 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
|
|
| 53 | c5visB_ |
|
not modelled |
43.1 |
17 |
PDB header:hydrolase,oxidoreductase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: 1.73 angstrom resolution crystal structure of dihydropteroate synthase2 (folp-smz_b27) from soil uncultured bacterium.
|
|
|
| 54 | c3nm3D_ |
|
not modelled |
39.1 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:thiamine biosynthetic bifunctional enzyme;
PDBTitle: the crystal structure of candida glabrata thi6, a bifunctional enzyme2 involved in thiamin biosyhthesis of eukaryotes
|
|
|
| 55 | c3ipwA_ |
|
not modelled |
37.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase tatd family protein;
PDBTitle: crystal structure of hydrolase tatd family protein from entamoeba2 histolytica
|
|
|
| 56 | c5uswD_ |
|
not modelled |
37.6 |
8 |
PDB header:transferase
Chain: D: PDB Molecule:dihydropteroate synthase;
PDBTitle: the crystal structure of 7,8-dihydropteroate synthase from vibrio2 fischeri es114
|
|
|
| 57 | c3f4cA_ |
|
not modelled |
35.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:organophosphorus hydrolase;
PDBTitle: crystal structure of organophosphorus hydrolase from geobacillus2 stearothermophilus strain 10, with glycerol bound
|
|
|
| 58 | c4djdD_ |
|
not modelled |
34.3 |
11 |
PDB header:transferase/vitamin-binding protein
Chain: D: PDB Molecule:corrinoid/iron-sulfur protein small subunit;
PDBTitle: crystal structure of folate-free corrinoid iron-sulfur protein (cfesp)2 in complex with its methyltransferase (metr)
|
|
|
| 59 | c6o5lA_ |
|
not modelled |
31.3 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ppra;
PDBTitle: crystal structure of ppra filament from deinococcus peraridilitoris
|
|
|
| 60 | c3ceuA_ |
|
not modelled |
29.8 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamine phosphate pyrophosphorylase (bt_0647)2 from bacteroides thetaiotaomicron. northeast structural genomics3 consortium target btr268
|
|
|
| 61 | c2d16B_ |
|
not modelled |
29.0 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ph1918;
PDBTitle: crystal structure of ph1918 protein from pyrococcus horikoshii ot3
|
|
|
| 62 | d1gkma_ |
|
not modelled |
28.9 |
18 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:HAL/PAL-like |
|
|
| 63 | c2h9aB_ |
|
not modelled |
28.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-sulfur protein;
PDBTitle: corrinoid iron-sulfur protein
|
|
|
| 64 | d1vgga_ |
|
not modelled |
26.5 |
27 |
Fold:Ta1353-like
Superfamily:Ta1353-like
Family:Ta1353-like |
|
|
| 65 | c2ekmC_ |
|
not modelled |
24.2 |
17 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein st1511;
PDBTitle: structure of st1219 protein from sulfolobus tokodaii
|
|
|
| 66 | c3unvB_ |
|
not modelled |
24.2 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:admh;
PDBTitle: pantoea agglomerans phenylalanine aminomutase
|
|
|
| 67 | c6s7qG_ |
|
not modelled |
23.7 |
18 |
PDB header:lyase
Chain: G: PDB Molecule:ergothionase;
PDBTitle: crystal structure of ergothioneine degrading enzyme ergothionase from2 treponema denticola in complex with desmethyl-ergothioneine sulfonic3 acid
|
|
|
| 68 | c6cluC_ |
|
not modelled |
21.4 |
14 |
PDB header:antimicrobial protein
Chain: C: PDB Molecule:dihydropteroate synthase;
PDBTitle: staphylococcus aureus dihydropteroate synthase (sadhps) f17l e208k2 double mutant structure
|
|
|
| 69 | c2vefB_ |
|
not modelled |
21.2 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
|
|
| 70 | c2o6yF_ |
|
not modelled |
20.7 |
22 |
PDB header:lyase
Chain: F: PDB Molecule:putative histidine ammonia-lyase;
PDBTitle: tyrosine ammonia-lyase from rhodobacter sphaeroides
|
|
|
| 71 | d2flia1 |
|
not modelled |
20.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 72 | c2qveA_ |
|
not modelled |
20.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:tyrosine aminomutase;
PDBTitle: crystal structure of sgtam bound to mechanism based inhibitor
|
|
|
| 73 | d3bofa1 |
|
not modelled |
19.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
|
|
| 74 | d1rd5a_ |
|
not modelled |
19.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 75 | c3k5pA_ |
|
not modelled |
19.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
|
|
| 76 | d1dosa_ |
|
not modelled |
18.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 77 | c6mc8A_ |
|
not modelled |
17.7 |
30 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna repair protein ppra;
PDBTitle: crystal structure of ppra dimer from deinococcus deserti
|
|
|
| 78 | c4exaD_ |
|
not modelled |
17.4 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the pa4992, the putative aldo-keto reductase from2 pseudomona aeruginosa
|
|
|
| 79 | d1ur4a_ |
|
not modelled |
17.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 80 | c3i5aA_ |
|
not modelled |
17.0 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator/ggdef domain protein;
PDBTitle: crystal structure of full-length wpsr from pseudomonas syringae
|
|
|
| 81 | c3uowB_ |
|
not modelled |
15.8 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:gmp synthetase;
PDBTitle: crystal structure of pf10_0123, a gmp synthetase from plasmodium2 falciparum
|
|
|
| 82 | c5n2pA_ |
|
not modelled |
15.7 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 83 | c2gl0A_ |
|
not modelled |
14.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: structure of pae2307 in complex with adenosine
|
|
|
| 84 | c2eklA_ |
|
not modelled |
14.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|
|
|
| 85 | d1o9ga_ |
|
not modelled |
14.0 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA methyltransferase AviRa |
|
|
| 86 | c3pm6B_ |
|
not modelled |
13.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
|
|
| 87 | c4muzA_ |
|
not modelled |
13.7 |
16 |
PDB header:lyase/lyase inhibitor
Chain: A: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 archaeoglobus fulgidus complexed with inhibitor bmp
|
|
|
| 88 | c3exsB_ |
|
not modelled |
13.7 |
6 |
PDB header:lyase
Chain: B: PDB Molecule:rmpd (hexulose-6-phosphate synthase);
PDBTitle: crystal structure of kgpdc from streptococcus mutans in2 complex with d-r5p
|
|
|
| 89 | c5yf1A_ |
|
not modelled |
12.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:carnosine n-methyltransferase;
PDBTitle: crystal structure of carnmt1 bound to carnosine and sfg
|
|
|
| 90 | c3ufbA_ |
|
not modelled |
12.7 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:type i restriction-modification system methyltransferase
PDBTitle: crystal structure of a modification subunit of a putative type i2 restriction enzyme from vibrio vulnificus yj016
|
|
|
| 91 | d1gvea_ |
|
not modelled |
11.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:NAD(P)-linked oxidoreductase
Family:Aldo-keto reductases (NADP) |
|
|
| 92 | c3jr2D_ |
|
not modelled |
11.7 |
9 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:hexulose-6-phosphate synthase sgbh;
PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 93 | c2rdtA_ |
|
not modelled |
10.6 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hydroxyacid oxidase 1;
PDBTitle: crystal structure of human glycolate oxidase (go) in complex with cdst
|
|
|
| 94 | c4o1fB_ |
|
not modelled |
10.1 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase dhps;
PDBTitle: structure of a methyltransferase component in complex with thf2 involved in o-demethylation
|
|
|
| 95 | c5tw7E_ |
|
not modelled |
9.0 |
23 |
PDB header:ligase
Chain: E: PDB Molecule:gmp synthase [glutamine-hydrolyzing];
PDBTitle: crystal structure of a gmp synthase (glutamine-hydrolyzing) from2 neisseria gonorrhoeae
|
|
|
| 96 | c1gpmD_ |
|
not modelled |
9.0 |
21 |
PDB header:transferase (glutamine amidotransferase)
Chain: D: PDB Molecule:gmp synthetase;
PDBTitle: escherichia coli gmp synthetase complexed with amp and pyrophosphate
|
|
|
| 97 | d2q02a1 |
|
not modelled |
9.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 98 | c5z79F_ |
|
not modelled |
8.8 |
15 |
PDB header:transferase
Chain: F: PDB Molecule:hydroxymethyldihydropterin pyrophosphokinase-
PDBTitle: crystal structure analysis of the hppk-dhps in complex with substrates
|
|
|
| 99 | c5ybbA_ |
|
not modelled |
8.8 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:type i restriction-modification system methyltransferase
PDBTitle: structural basis underlying complex assembly andconformational2 transition of the type i r-m system
|
|
|