| 1 |
|
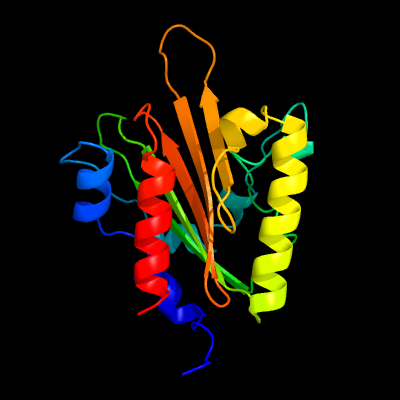
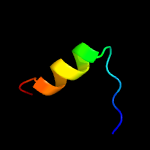
PDB 4esq chain A
Region: 33 - 209
Aligned: 172
Modelled: 177
Confidence: 100.0%
Identity: 19%
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase;
PDBTitle: crystal structure of the extracellular domain of pknh from2 mycobacterium tuberculosis
Phyre2
| 2 |
|
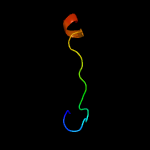
PDB 4ash chain B
Region: 146 - 186
Aligned: 41
Modelled: 41
Confidence: 48.6%
Identity: 27%
PDB header:hydrolase
Chain: B: PDB Molecule:ns6 protease;
PDBTitle: crystal structure of the ns6 protease from murine norovirus 1
Phyre2
| 3 |
|
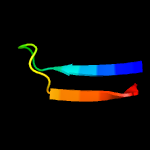
PDB 3lo3 chain E
Region: 121 - 139
Aligned: 19
Modelled: 19
Confidence: 22.6%
Identity: 5%
PDB header:structure genomics, unknown function
Chain: E: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 colwellia psychrerythraea 34h.
Phyre2
| 4 |
|
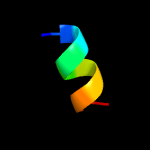
PDB 1pft chain A
Region: 84 - 104
Aligned: 21
Modelled: 21
Confidence: 15.4%
Identity: 29%
Fold: Rubredoxin-like
Superfamily: Zinc beta-ribbon
Family: Transcriptional factor domain
Phyre2
| 5 |
|
PDB 2fiu chain A domain 1
Region: 121 - 139
Aligned: 19
Modelled: 19
Confidence: 15.0%
Identity: 5%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: Atu0297-like
Phyre2
| 6 |
|
PDB 1dl6 chain A
Region: 84 - 104
Aligned: 21
Modelled: 21
Confidence: 13.9%
Identity: 29%
Fold: Rubredoxin-like
Superfamily: Zinc beta-ribbon
Family: Transcriptional factor domain
Phyre2
| 7 |
|
PDB 3k1f chain M
Region: 84 - 104
Aligned: 21
Modelled: 21
Confidence: 13.3%
Identity: 24%
PDB header:transcription
Chain: M: PDB Molecule:transcription initiation factor iib;
PDBTitle: crystal structure of rna polymerase ii in complex with tfiib
Phyre2
| 8 |
|
PDB 5a6w chain C
Region: 172 - 189
Aligned: 17
Modelled: 18
Confidence: 12.9%
Identity: 12%
PDB header:antiviral protein
Chain: C: PDB Molecule:avr-pik protein;
PDBTitle: complex of rice blast (magnaporthe oryzae) effector protein avr-pikd2 with the hma domain of pikp1 from rice (oryza sativa)
Phyre2
| 9 |
|
PDB 2js3 chain B
Region: 192 - 208
Aligned: 17
Modelled: 17
Confidence: 12.2%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of protein q6n9a4_rhopa. northeast structural genomics2 consortium target rpt8
Phyre2
| 10 |
|
PDB 4wj2 chain A
Region: 21 - 60
Aligned: 40
Modelled: 40
Confidence: 12.0%
Identity: 20%
PDB header:unknown function
Chain: A: PDB Molecule:antigen mtb48;
PDBTitle: mycobacterial protein
Phyre2
| 11 |
|
PDB 4i0x chain A
Region: 97 - 106
Aligned: 10
Modelled: 10
Confidence: 10.5%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:esat-6-like protein mab_3112;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 12 |
|
PDB 3dca chain C
Region: 107 - 139
Aligned: 33
Modelled: 31
Confidence: 10.1%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:rpa0582;
PDBTitle: crystal structure of the rpa0582- protein of unknown2 function from rhodopseudomonas palustris- a structural3 genomics target
Phyre2
| 13 |
|
PDB 2b3g chain B
Region: 32 - 53
Aligned: 19
Modelled: 22
Confidence: 9.4%
Identity: 11%
PDB header:replication
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53n (fragment 33-60) bound to rpa70n
Phyre2
| 14 |
|
PDB 4zjf chain A
Region: 166 - 193
Aligned: 28
Modelled: 28
Confidence: 8.2%
Identity: 18%
PDB header:viral protein
Chain: A: PDB Molecule:glycoprotein;
PDBTitle: crystal structure of gp1 - the receptor binding domain of lassa virus
Phyre2
| 15 |
|
PDB 3i08 chain D
Region: 125 - 141
Aligned: 17
Modelled: 17
Confidence: 7.8%
Identity: 18%
PDB header:signaling protein
Chain: D: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: crystal structure of the s1-cleaved notch1 negative2 regulatory region (nrr)
Phyre2
| 16 |
|
PDB 1hsk chain A
Region: 155 - 206
Aligned: 52
Modelled: 52
Confidence: 6.6%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of s. aureus murb
Phyre2
| 17 |
|
PDB 1zzp chain A
Region: 22 - 56
Aligned: 31
Modelled: 35
Confidence: 5.3%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:proto-oncogene tyrosine-protein kinase abl1;
PDBTitle: solution structure of the f-actin binding domain of bcr-2 abl/c-abl
Phyre2
| 18 |
|
PDB 2h8b chain A
Region: 12 - 23
Aligned: 12
Modelled: 12
Confidence: 5.0%
Identity: 17%
PDB header:hormone/growth factor
Chain: A: PDB Molecule:insulin-like 3;
PDBTitle: solution structure of insl3
Phyre2
| 19 |
|
PDB 2k6t chain A
Region: 12 - 23
Aligned: 12
Modelled: 12
Confidence: 5.0%
Identity: 17%
PDB header:hormone
Chain: A: PDB Molecule:insulin-like 3 a chain;
PDBTitle: solution structure of the relaxin-like factor
Phyre2