1 c3q4hB_
100.0
52
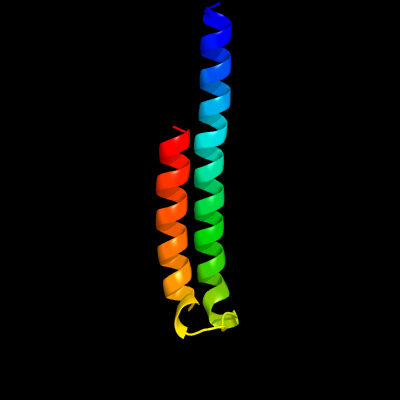
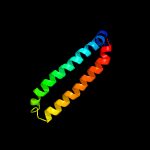
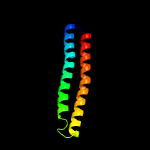
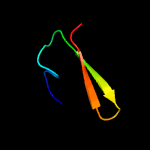
PDB header: metal transportChain: B: PDB Molecule: low molecular weight protein antigen 7;PDBTitle: crystal structure of the mycobacterium smegmatis esxgh complex2 (msmeg_0620-msmeg_0621)
2 c3h6pD_
100.0
65
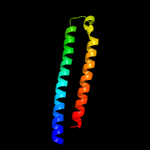
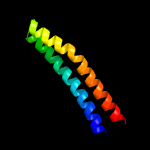
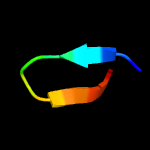
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: esat-6-like protein esxr;PDBTitle: crystal structure of rv3019c-rv3020c from mycobacterium tuberculosis
3 c2kg7B_
98.6
56
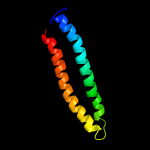
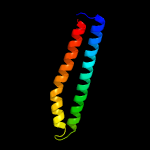
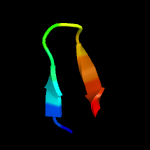
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
4 d1wa8b1
98.4
18
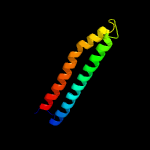
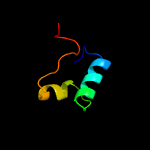
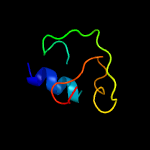
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
5 c4lwsB_
97.5
12
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
6 c4lwsA_
97.1
15
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
7 d1wa8a1
95.6
13
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
8 c3gvmA_
95.4
9
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c4iogD_
94.5
14
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
10 c3zbhC_
90.4
9
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
11 c2vs0B_
90.0
11
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
12 c2do5A_
29.6
45
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: splicing factor 3b subunit 2;PDBTitle: solution structure of the sap domain of human splicing2 factor 3b subunit 2
13 d1ju8a_
26.3
35
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Albumin 1
14 c3wofB_
25.9
60
PDB header: transferase/transcriptionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of p23-45 gp39 (6-132) bound to thermus thermophilus2 rna polymerase beta-flap domain
15 c3wodG_
23.2
60
PDB header: transferase/transcriptionChain: G: PDB Molecule: putative uncharacterized protein;PDBTitle: rna polymerase-gp39 complex
16 c5tc1M_
21.4
46
PDB header: viral protein/rnaChain: M: PDB Molecule: maturation protein;PDBTitle: in situ structures of the genome and genome-delivery apparatus in2 ssrna bacteriophage ms2
17 c3qbzA_
21.2
33
PDB header: cell cycleChain: A: PDB Molecule: ddk kinase regulatory subunit dbf4;PDBTitle: crystal structure of the rad53-recognition domain of saccharomyces2 cerevisiae dbf4
18 c5t2sA_
20.0
25
PDB header: cell cycleChain: A: PDB Molecule: ddk kinase regulatory subunit dbf4,serine/threonine-proteinPDBTitle: structure of the fha1 domain of rad53 bound simultaneously to the brct2 domain of dbf4 and a phosphopeptide.
19 d1p8ba_
19.8
35
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Albumin 1
20 d3cr3a1
19.6
23
Fold: DhaL-likeSuperfamily: DhaL-likeFamily: DhaL-like
21 c2ru8A_
not modelled
16.3
35
PDB header: replicationChain: A: PDB Molecule: primosomal protein 1;PDBTitle: dnat c-terminal domain
22 c6d8tA_
not modelled
14.6
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant e25k/k27e
23 d1scya_
not modelled
14.5
33
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
24 c1scyA_
not modelled
14.5
33
PDB header: neurotoxinChain: A: PDB Molecule: scyllatoxin;PDBTitle: determination of the three-dimensional structure of scyllatoxin by 1h2 nuclear magnetic resonance
25 c5is0E_
not modelled
14.2
21
PDB header: transport proteinChain: E: PDB Molecule: transient receptor potential cation channel subfamily vPDBTitle: structure of trpv1 in complex with capsazepine, determined in lipid2 nanodisc
26 c2izpB_
not modelled
13.6
7
PDB header: toxinChain: B: PDB Molecule: putative membrane antigen;PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.
27 c2ky3A_
not modelled
13.1
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: solution structure of gs-alfa-ktx5.4 synthetic scorpion like
28 c6d93A_
not modelled
13.0
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant y31a
29 d1un8a1
not modelled
12.8
17
Fold: DhaL-likeSuperfamily: DhaL-likeFamily: DhaL-like
30 c4c0lA_
not modelled
12.5
17
PDB header: hydrolaseChain: A: PDB Molecule: mitochondrial rho gtpase;PDBTitle: crystal structure of drosophila miro ef hand and cgtpase domains2 bound to one magnesium ion and mg:gdp (mggdp-miros)
31 c6d8rA_
not modelled
12.5
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant e25k
32 c6d8yA_
not modelled
12.3
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant y31h
33 d2izpa1
not modelled
11.3
7
Fold: IpaD-likeSuperfamily: IpaD-likeFamily: IpaD-like
34 c4bjjB_
not modelled
11.3
54
PDB header: transcriptionChain: B: PDB Molecule: transcription factor tau subunit sfc7;PDBTitle: sfc1-sfc7 dimerization module
35 c6d8qA_
not modelled
11.0
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant dp30/y31+n
36 c4o8uE_
not modelled
10.9
53
PDB header: unknown functionChain: E: PDB Molecule: uncharacterized protein pf2046;PDBTitle: structure of pf2046
37 c6d3tA_
not modelled
10.8
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant dp30
38 d1pnha_
not modelled
10.8
33
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
39 c1pnhA_
not modelled
10.8
33
PDB header: toxinChain: A: PDB Molecule: scorpion toxin;PDBTitle: solution structure of po5-nh2, a scorpion toxin analog with high2 affinity for the apamin-sensitive potassium channel
40 c3zqbB_
not modelled
10.7
25
PDB header: cell invasionChain: B: PDB Molecule: protein prgi, cell invasion protein sipd;PDBTitle: prgi-sipd from salmonella typhimurium
41 c6d9oA_
not modelled
10.6
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant e25a
42 c6d8sA_
not modelled
10.2
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant k27e
43 c2p7nA_
not modelled
9.6
31
PDB header: cell invasionChain: A: PDB Molecule: pathogenicity island 1 effector protein;PDBTitle: crystal structure of the pathogenicity island 1 effector protein from2 chromobacterium violaceum. northeast structural genomics consortium3 (nesgc) target cvr69.
44 c6d9pA_
not modelled
9.6
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant k27a
45 c2iunD_
not modelled
9.0
67
PDB header: viral proteinChain: D: PDB Molecule: avian adenovirus celo long fibre;PDBTitle: structure of the c-terminal head domain of the avian adenovirus celo2 long fibre (p21 crystal form)
46 c6d8uA_
not modelled
8.7
35
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant k20e
47 d1e6va1
not modelled
8.7
38
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
48 c4hdeA_
not modelled
8.0
16
PDB header: lipid binding proteinChain: A: PDB Molecule: sco1/senc family lipoprotein;PDBTitle: the crystal structure of a sco1/senc family lipoprotein from bacillus2 anthracis str. ames
49 c3nzzA_
not modelled
7.9
25
PDB header: cell invasionChain: A: PDB Molecule: cell invasion protein sipd;PDBTitle: crystal structure of the salmonella type iii secretion system tip2 protein sipd
50 c4b2gB_
not modelled
7.0
34
PDB header: signaling proteinChain: B: PDB Molecule: gh3-1 auxin conjugating enzyme;PDBTitle: crystal structure of an indole-3-acetic acid amido synthase from vitis2 vinifera involved in auxin homeostasis
51 d1aisa1
not modelled
6.8
30
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain
52 d1f15a_
not modelled
6.7
42
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP
53 d1r5ta_
not modelled
6.7
23
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Cytidine deaminase
54 c3dktD_
not modelled
6.6
18
PDB header: structural protein/virus like particleChain: D: PDB Molecule: maritimacin;PDBTitle: crystal structure of thermotoga maritima encapsulin
55 c6c3rB_
not modelled
6.6
46
PDB header: viral proteinChain: B: PDB Molecule: cricket paralysis virus 1a protein;PDBTitle: cricket paralysis virus rnai suppressor protein crpv-1a
56 c3fybA_
not modelled
6.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein of unknown function (duf1244);PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
57 c6avhA_
not modelled
6.1
24
PDB header: ligase, plant proteinChain: A: PDB Molecule: gh3.15 acyl acid amido synthetase;PDBTitle: gh3.15 acyl acid amido synthetase
58 c5n1qD_
not modelled
6.0
45
PDB header: transferaseChain: D: PDB Molecule: methyl-coenzyme m reductase iii from methanothermococcusPDBTitle: methyl-coenzyme m reductase iii from methanothermococcus2 thermolithotrophicus at 1.9 a resolution
59 c1mtpB_
not modelled
6.0
50
PDB header: structural genomicsChain: B: PDB Molecule: serine proteinase inhibitor (serpin), chain b;PDBTitle: the x-ray crystal structure of a serpin from a thermophilic prokaryote
60 d1laja_
not modelled
6.0
24
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Bromoviridae-like VP
61 c1lajA_
not modelled
6.0
24
PDB header: virus/rnaChain: A: PDB Molecule: capsid protein;PDBTitle: the structure of tomato aspermy virus by x-ray2 crystallography
62 d2o35a1
not modelled
5.9
22
Fold: SMc04008-likeSuperfamily: SMc04008-likeFamily: SMc04008-like
63 c2o35A_
not modelled
5.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf1244;PDBTitle: protein of unknown function (duf1244) from sinorhizobium meliloti
64 d2akab1
not modelled
5.8
35
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins
65 c3jcuO_
not modelled
5.8
44
PDB header: membrane proteinChain: O: PDB Molecule: oxygen-evolving enhancer protein 1, chloroplastic;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
66 c2jorA_
not modelled
5.7
38
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha chain;PDBTitle: nmr solution structure, stability, and interaction of the2 recombinant bovine fibrinogen alphac-domain fragment
67 d1hbna1
not modelled
5.7
40
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
68 d1vi7a1
not modelled
5.5
35
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: YigZ N-terminal domain-like
69 d1e6ya1
not modelled
5.5
45
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainSuperfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domainFamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
70 d2axto1
not modelled
5.4
40
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: PsbO-like
71 c3g8aF_
not modelled
5.4
14
PDB header: transferaseChain: F: PDB Molecule: ribosomal rna small subunit methyltransferase g;PDBTitle: t. thermophilus 16s rrna g527 methyltransferase in complex with adohcy2 in space group p61
72 c5lnkm_
not modelled
5.3
38
PDB header: oxidoreductaseChain: M: PDB Molecule: mitochondrial complex i, nd4 subunit;PDBTitle: entire ovine respiratory complex i