| 1 |
|
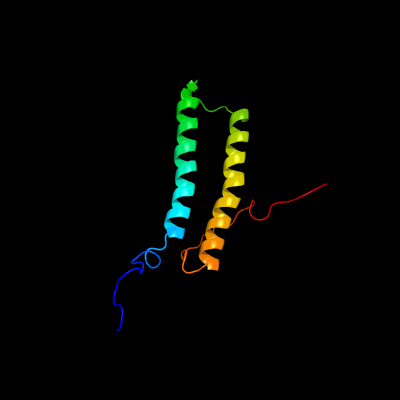
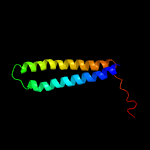
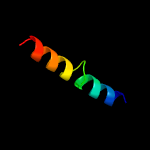
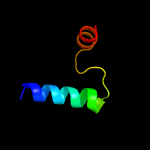
PDB 2kg7 chain A
Region: 1 - 97
Aligned: 97
Modelled: 97
Confidence: 100.0%
Identity: 92%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein esxg (pe family protein);
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 2 |
|
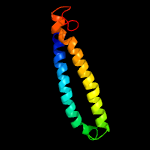
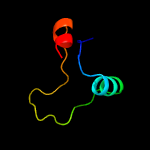
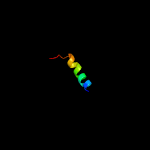
PDB 3h6p chain B
Region: 14 - 80
Aligned: 67
Modelled: 67
Confidence: 100.0%
Identity: 100%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:esat-6 like protein esxs;
PDBTitle: crystal structure of rv3019c-rv3020c from mycobacterium tuberculosis
Phyre2
| 3 |
|
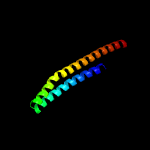
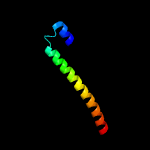
PDB 2kg7 chain B
Region: 1 - 96
Aligned: 96
Modelled: 96
Confidence: 95.7%
Identity: 18%
PDB header:unknown function
Chain: B: PDB Molecule:esat-6-like protein esxh;
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 4 |
|
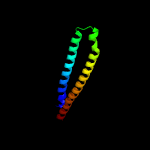
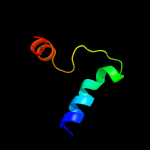
PDB 1wa8 chain A domain 1
Region: 10 - 93
Aligned: 84
Modelled: 84
Confidence: 95.3%
Identity: 30%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 5 |
|
PDB 1wa8 chain B domain 1
Region: 9 - 94
Aligned: 86
Modelled: 86
Confidence: 95.3%
Identity: 16%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 6 |
|
PDB 3zbh chain C
Region: 10 - 96
Aligned: 87
Modelled: 87
Confidence: 93.9%
Identity: 20%
PDB header:unknown function
Chain: C: PDB Molecule:esxa;
PDBTitle: geobacillus thermodenitrificans esxa crystal form i
Phyre2
| 7 |
|
PDB 4lws chain A
Region: 9 - 93
Aligned: 85
Modelled: 84
Confidence: 93.1%
Identity: 21%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 8 |
|
PDB 3gvm chain A
Region: 1 - 93
Aligned: 93
Modelled: 93
Confidence: 90.3%
Identity: 17%
PDB header:viral protein
Chain: A: PDB Molecule:putative uncharacterized protein sag1039;
PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
Phyre2
| 9 |
|
PDB 4lws chain B
Region: 9 - 83
Aligned: 75
Modelled: 75
Confidence: 88.2%
Identity: 17%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 10 |
|
PDB 2vs0 chain B
Region: 10 - 87
Aligned: 78
Modelled: 78
Confidence: 74.2%
Identity: 14%
PDB header:cell invasion
Chain: B: PDB Molecule:virulence factor esxa;
PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
Phyre2
| 11 |
|
PDB 4iog chain D
Region: 1 - 86
Aligned: 86
Modelled: 86
Confidence: 66.9%
Identity: 19%
PDB header:unknown function
Chain: D: PDB Molecule:secreted protein esxb;
PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
Phyre2
| 12 |
|
PDB 2v0x chain B
Region: 12 - 38
Aligned: 25
Modelled: 27
Confidence: 11.2%
Identity: 52%
PDB header:cell cycle
Chain: B: PDB Molecule:lamina-associated polypeptide 2 isoforms alpha/zeta;
PDBTitle: the dimerization domain of lap2alpha
Phyre2
| 13 |
|
PDB 2q6m chain A
Region: 51 - 94
Aligned: 44
Modelled: 44
Confidence: 9.3%
Identity: 30%
PDB header:toxin
Chain: A: PDB Molecule:cholix toxin;
PDBTitle: catalytic fragment of cholix toxin from vibrio cholerae in complex2 with the pj34 inhibitor
Phyre2
| 14 |
|
PDB 1xrx chain D
Region: 43 - 48
Aligned: 6
Modelled: 6
Confidence: 7.9%
Identity: 83%
PDB header:replication inhibitor
Chain: D: PDB Molecule:seqa protein;
PDBTitle: crystal structure of a dna-binding protein
Phyre2
| 15 |
|
PDB 1xrx chain A domain 1
Region: 43 - 48
Aligned: 6
Modelled: 6
Confidence: 7.9%
Identity: 83%
Fold: Ribbon-helix-helix
Superfamily: Ribbon-helix-helix
Family: SeqA N-terminal domain-like
Phyre2
| 16 |
|
PDB 3u0c chain A
Region: 24 - 73
Aligned: 50
Modelled: 50
Confidence: 6.8%
Identity: 18%
PDB header:cell invasion
Chain: A: PDB Molecule:invasin ipab;
PDBTitle: crystal structure of n-terminal region of type iii secretion first2 translocator ipab (residues 74-224)
Phyre2
| 17 |
|
PDB 1hbn chain A domain 1
Region: 43 - 75
Aligned: 32
Modelled: 33
Confidence: 6.7%
Identity: 44%
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Superfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Family: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Phyre2
| 18 |
|
PDB 5v6h chain C
Region: 59 - 70
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 42%
PDB header:protein binding
Chain: C: PDB Molecule:pdz domain-containing protein gipc2;
PDBTitle: crystal structure of myosin vi in complex with gh2 domain of gipc2
Phyre2
| 19 |
|
PDB 1e6v chain A domain 1
Region: 43 - 75
Aligned: 32
Modelled: 33
Confidence: 5.7%
Identity: 38%
Fold: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Superfamily: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Family: Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Phyre2
| 20 |
|
PDB 4e1r chain A
Region: 46 - 65
Aligned: 20
Modelled: 20
Confidence: 5.4%
Identity: 30%
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: crystal structure of the dimerization domain of lsr2 from2 mycobacterium tuberculosis in the p 31 2 1 space group
Phyre2
| 21 |
|
| 22 |
|