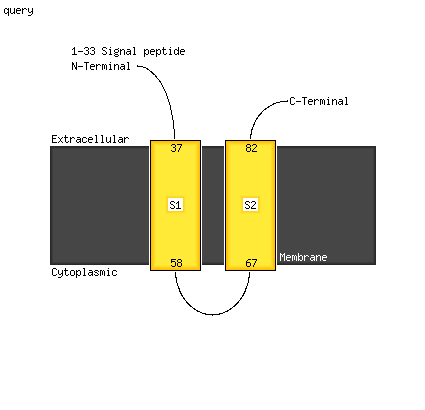
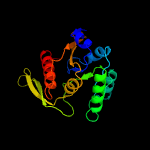
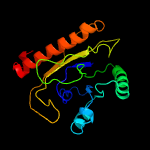
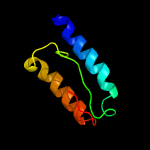
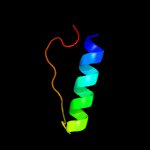
1 c5kymA_
100.0
18
PDB header: transferaseChain: A: PDB Molecule: 1-acyl-sn-glycerol-3-phosphate acyltransferase;PDBTitle: crystal structure of the 1-acyl-sn-glycerophosphate (lpa)2 acyltransferase, plsc, from thermotoga maritima
2 c5f34A_
99.3
12
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol mannoside acyltransferase;PDBTitle: crystal structure of membrane associated pata from mycobacterium2 smegmatis in complex with s-hexadecyl coenzyme a - p21 space group
3 c5knkB_
99.0
13
PDB header: transferaseChain: B: PDB Molecule: lipid a biosynthesis lauroyl acyltransferase;PDBTitle: lipid a secondary acyltransferase lpxm from acinetobacter baumannii2 with catalytic residue substitution (e127a)
4 d1iuqa_
97.9
21
Fold: Glycerol-3-phosphate (1)-acyltransferaseSuperfamily: Glycerol-3-phosphate (1)-acyltransferaseFamily: Glycerol-3-phosphate (1)-acyltransferase
5 c5vrhA_
78.5
14
PDB header: transferaseChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
6 c4hg3C_
49.0
24
PDB header: hydrolaseChain: C: PDB Molecule: probable hydrolase nit2;PDBTitle: structural insights into yeast nit2: wild-type yeast nit2 in complex2 with alpha-ketoglutarate
7 c4cyyA_
43.2
17
PDB header: hydrolaseChain: A: PDB Molecule: pantetheinase;PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
8 c2e11B_
38.5
10
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
9 c3wuyA_
36.3
28
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase;PDBTitle: crystal structure of nit6803
10 c3hkxA_
33.0
24
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
11 c4mxnB_
30.5
13
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative glycosyl hydrolase (parmer_00599) from2 parabacteroides merdae atcc 43184 at 1.95 a resolution
12 c1emsB_
28.4
11
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
13 c2vhhA_
27.0
21
PDB header: hydrolaseChain: A: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
14 c1yj5B_
26.9
22
PDB header: transferaseChain: B: PDB Molecule: 5' polynucleotide kinase-3' phosphatase catalytic domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
15 c6i00C_
26.3
20
PDB header: hydrolaseChain: C: PDB Molecule: bifunctional nitrilase/nitrile hydratase nit4;PDBTitle: cryo-em informed directed evolution of nitrilase 4 leads to a change2 in quaternary structure.
16 d1k7ja_
23.4
16
Fold: YrdC/RibBSuperfamily: YrdC/RibBFamily: YrdC-like
17 c3zvmA_
22.1
23
PDB header: hydrolase/transferase/dnaChain: A: PDB Molecule: bifunctional polynucleotide phosphatase/kinase;PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
18 c6mg6D_
21.8
9
PDB header: hydrolaseChain: D: PDB Molecule: carbon-nitrogen hydrolase;PDBTitle: crystal structure of carbon-nitrogen hydrolase from helicobacter2 pylori g27
19 c6ftqA_
21.0
11
PDB header: hydrolaseChain: A: PDB Molecule: beta-ureidopropionase;PDBTitle: crystal structure of human beta-ureidopropionase (beta-alanine2 synthase) - mutant t299c
20 d1uf5a_
20.8
14
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
21 d1emsa2
not modelled
19.1
14
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
22 c5uyvA_
not modelled
18.6
19
PDB header: metal transportChain: A: PDB Molecule: periplasmic chelated iron-binding protein yfea;PDBTitle: yfea ancillary sites that do not co-load with site 2
23 c2vhiG_
not modelled
18.0
17
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
24 d1hrua_
not modelled
17.7
17
Fold: YrdC/RibBSuperfamily: YrdC/RibBFamily: YrdC-like
25 c2o1eB_
not modelled
17.6
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ycdh;PDBTitle: crystal structure of the metal-dependent lipoprotein ycdh2 from bacillus subtilis, northeast structural genomics3 target sr583
26 d1vj0a1
not modelled
16.7
27
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain
27 c3ilvA_
not modelled
16.3
20
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
28 c5khaA_
not modelled
15.3
8
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad+ synthetase;PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
29 c2w1vA_
not modelled
15.3
17
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
30 d1j31a_
not modelled
14.9
20
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
31 d1jcua_
not modelled
14.6
26
Fold: YrdC/RibBSuperfamily: YrdC/RibBFamily: YrdC-like
32 c5ixdB_
not modelled
14.1
19
PDB header: cytokineChain: B: PDB Molecule: interferon lambda receptor 1;PDBTitle: structure of human jak1 ferm/sh2 in complex with ifn lambda receptor
33 c4makA_
not modelled
13.1
16
PDB header: hydrolaseChain: A: PDB Molecule: crispr-associated endoribonuclease cas2;PDBTitle: crystal structure of a putative ssrna endonuclease cas2, crispr2 adaptation protein from e.coli
34 c3hjtB_
not modelled
12.0
16
PDB header: cell adhesion, transport proteinChain: B: PDB Molecule: lmb;PDBTitle: structure of laminin binding protein (lmb) of streptococcus agalactiae2 a bifunctional protein with adhesin and metal transporting activity
35 d1bg5a1
not modelled
12.0
11
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
36 c2ov3A_
not modelled
11.5
15
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein component of an abc type zincPDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc bound
37 d1f89a_
not modelled
11.4
14
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
38 d1pq4a_
not modelled
11.1
13
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
39 d1bhea_
not modelled
10.9
14
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase
40 c3zppA_
not modelled
10.4
30
PDB header: cell adhesionChain: A: PDB Molecule: cell wall surface anchor family protein;PDBTitle: structure of the streptococcus pneumoniae surface protein2 and adhesin pfba
41 c5h8lM_
not modelled
10.2
6
PDB header: hydrolaseChain: M: PDB Molecule: n-carbamoylputrescine amidohydrolase;PDBTitle: crystal structure of medicago truncatula n-carbamoylputrescine2 amidohydrolase (mtcpa) c158s mutant in complex with putrescine
42 c4cl2A_
not modelled
10.1
17
PDB header: transport proteinChain: A: PDB Molecule: periplasmic solute binding protein;PDBTitle: structure of periplasmic metal binding protein from candidatus2 liberibacter asiaticus
43 c4f4hA_
not modelled
9.9
17
PDB header: ligaseChain: A: PDB Molecule: glutamine dependent nad+ synthetase;PDBTitle: crystal structure of a glutamine dependent nad+ synthetase from2 burkholderia thailandensis
44 c3n05B_
not modelled
9.7
21
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
45 c2ogwB_
not modelled
9.3
16
PDB header: transport proteinChain: B: PDB Molecule: high-affinity zinc uptake system protein znuaPDBTitle: structure of abc type zinc transporter from e. coli
46 c3dlaD_
not modelled
8.9
24
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
47 c6f87C_
not modelled
8.8
35
PDB header: transferaseChain: C: PDB Molecule: threonylcarbamoyl-amp synthase;PDBTitle: crystal structure of p. abyssi sua5 complexed with l-threonine and ppi
48 c4c2lA_
not modelled
8.0
22
PDB header: hydrolaseChain: A: PDB Molecule: endo-xylogalacturonan hydrolase a;PDBTitle: crystal structure of endo-xylogalacturonan hydrolase from aspergillus2 tubingensis
49 d1gnea1
not modelled
8.0
11
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
50 d1b8xa1
not modelled
8.0
11
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
51 d1y0na_
not modelled
7.8
25
Fold: YehU-likeSuperfamily: YehU-likeFamily: YehU-like
52 c2xc8B_
not modelled
7.5
50
PDB header: viral proteinChain: B: PDB Molecule: gene 22 product;PDBTitle: crystal structure of the gene 22 product of the bacillus subtilis spp12 phage
53 c4as2D_
not modelled
7.3
13
PDB header: hydrolaseChain: D: PDB Molecule: phosphorylcholine phosphatase;PDBTitle: pseudomonas aeruginosa phosphorylcholine phosphatase. monoclinic form
54 c3jurA_
not modelled
7.3
22
PDB header: hydrolaseChain: A: PDB Molecule: exo-poly-alpha-d-galacturonosidase;PDBTitle: the crystal structure of a hyperthermoactive exopolygalacturonase from2 thermotoga maritima
55 c3eqnB_
not modelled
7.1
55
PDB header: hydrolaseChain: B: PDB Molecule: glucan 1,3-beta-glucosidase;PDBTitle: crystal structure of beta-1,3-glucanase from phanerochaete2 chrysosporium (lam55a)
56 c2eqaA_
not modelled
7.0
35
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein st1526;PDBTitle: crystal structure of the hypothetical sua5 protein from2 sulfolobus tokodaii
57 d1g26a_
not modelled
7.0
50
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Granulin repeatFamily: Granulin repeat
58 c4z42A_
not modelled
6.6
24
PDB header: hydrolaseChain: A: PDB Molecule: urease subunit gamma;PDBTitle: crystal structure of urease from yersinia enterocolitica
59 d1k3ya1
not modelled
6.5
8
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
60 d1m5ha2
not modelled
6.5
14
Fold: Ferredoxin-likeSuperfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferaseFamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
61 c5n6mA_
not modelled
6.0
14
PDB header: membrane proteinChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
62 c2ps3A_
not modelled
5.9
16
PDB header: metal transportChain: A: PDB Molecule: high-affinity zinc uptake system protein znua;PDBTitle: structure and metal binding properties of znua, a periplasmic zinc2 transporter from escherichia coli
63 c4chmB_
not modelled
5.8
32
PDB header: cell cycleChain: B: PDB Molecule: imc sub-compartment protein isp1;PDBTitle: structure of inner membrane complex (imc) sub-compartment protein 12 (isp1) from toxoplasma gondii
64 d1rmga_
not modelled
5.7
14
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Galacturonase
65 c2plqA_
not modelled
5.4
18
PDB header: hydrolaseChain: A: PDB Molecule: aliphatic amidase;PDBTitle: crystal structure of the amidase from geobacillus pallidus rapc8
66 c4furD_
not modelled
5.2
28
PDB header: hydrolaseChain: D: PDB Molecule: urease subunit gamma 2;PDBTitle: crystal structure of urease subunit gamma 2 from brucella melitensis2 biovar abortus 2308
67 d1u1ia1
not modelled
5.2
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain