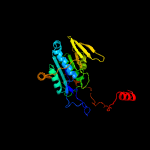
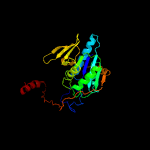
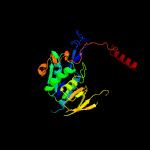
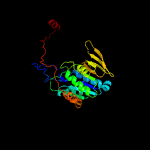
1 c6fahB_
100.0
34
PDB header: flavoproteinChain: B: PDB Molecule: caffeyl-coa reductase-etf complex subunit card;PDBTitle: molecular basis of the flavin-based electron-bifurcating caffeyl-coa2 reductase reaction
2 c5ol2E_
100.0
34
PDB header: flavoproteinChain: E: PDB Molecule: electron transfer flavoprotein small subunit;PDBTitle: the electron transferring flavoprotein/butyryl-coa dehydrogenase2 complex from clostridium difficile
3 c4kpuB_
100.0
33
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein alpha/beta-subunit;PDBTitle: electron transferring flavoprotein of acidaminococcus fermentans:2 towards a mechanism of flavin-based electron bifurcation
4 d1efvb_
100.0
30
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
5 c5ow0B_
100.0
28
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein, beta subunit;PDBTitle: crystal structure of an electron transfer flavoprotein from2 geobacillus metallireducens
6 d3clsc1
100.0
29
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
7 d1efpb_
100.0
33
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
8 d1o94c_
100.0
31
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
9 c1o94D_
99.9
15
PDB header: electron transportChain: D: PDB Molecule: electron transfer flavoprotein alpha-subunit;PDBTitle: ternary complex between trimethylamine dehydrogenase and2 electron transferring flavoprotein
10 d3clsd1
99.9
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
11 c3clrD_
99.9
16
PDB header: electron transportChain: D: PDB Molecule: electron transfer flavoprotein subunit alpha;PDBTitle: crystal structure of the r236a etf mutant from m. methylotrophus
12 c3ih5A_
99.9
16
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein alpha-subunit;PDBTitle: crystal structure of electron transfer flavoprotein alpha-subunit from2 bacteroides thetaiotaomicron
13 c4l2iA_
99.9
17
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein alpha subunit;PDBTitle: electron transferring flavoprotein of acidaminococcus fermentans:2 towards a mechanism of flavin-based electron bifurcation
14 c5ol2D_
99.9
11
PDB header: flavoproteinChain: D: PDB Molecule: electron transfer flavoprotein large subunit;PDBTitle: the electron transferring flavoprotein/butyryl-coa dehydrogenase2 complex from clostridium difficile
15 c1efvA_
99.8
17
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein;PDBTitle: three-dimensional structure of human electron transfer2 flavoprotein to 2.1 a resolution
16 c6fahE_
99.8
15
PDB header: flavoproteinChain: E: PDB Molecule: caffeyl-coa reductase-etf complex subunit care;PDBTitle: molecular basis of the flavin-based electron-bifurcating caffeyl-coa2 reductase reaction
17 d1efva1
99.8
17
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
18 c1t9gR_
99.7
17
PDB header: oxidoreductase, electron transportChain: R: PDB Molecule: electron transfer flavoprotein alpha-subunit,PDBTitle: structure of the human mcad:etf complex
19 c5ow0A_
99.5
22
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein, alpha subunit;PDBTitle: crystal structure of an electron transfer flavoprotein from2 geobacillus metallireducens
20 d1efpa1
99.5
20
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
21 c1efpC_
not modelled
99.4
20
PDB header: electron transportChain: C: PDB Molecule: protein (electron transfer flavoprotein);PDBTitle: electron transfer flavoprotein (etf) from paracoccus2 denitrificans
22 c3fetA_
not modelled
99.4
12
PDB header: electron transportChain: A: PDB Molecule: electron transfer flavoprotein subunit alpha relatedPDBTitle: crystal structure of the electron transfer flavoprotein subunit alpha2 related protein ta0212 from thermoplasma acidophilum
23 d1q77a_
not modelled
87.3
20
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
24 c4r2lB_
not modelled
86.5
15
PDB header: unknown functionChain: B: PDB Molecule: universal stress protein f;PDBTitle: crystal structure of ynaf (universal stress protein f) from salmonella2 typhimurium
25 c3fh0A_
not modelled
83.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative universal stress protein kpn_01444;PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
26 d2z3va1
not modelled
83.1
22
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
27 c4r2jA_
not modelled
82.7
14
PDB header: metal binding protein, unknown functionChain: A: PDB Molecule: universal stress protein e;PDBTitle: crystal structure of ydaa (universal stress protein e) from salmonella2 typhimurium
28 d1znna1
not modelled
82.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: PdxS-like
29 c2pfsA_
not modelled
81.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: universal stress protein;PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
30 c3mt0A_
not modelled
80.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pa1789;PDBTitle: the crystal structure of a functionally unknown protein pa1789 from2 pseudomonas aeruginosa pao1
31 c4wnyA_
not modelled
79.6
20
PDB header: signaling proteinChain: A: PDB Molecule: universal stress protein;PDBTitle: crystal structure of a protein from the universal stress protein2 family from burkholderia pseudomallei
32 c1znnF_
not modelled
79.6
19
PDB header: biosynthetic proteinChain: F: PDB Molecule: plp synthase;PDBTitle: structure of the synthase subunit of plp synthase
33 c2c4kD_
not modelled
79.6
12
PDB header: regulatory proteinChain: D: PDB Molecule: phosphoribosyl pyrophosphate synthetase-associated proteinPDBTitle: crystal structure of human phosphoribosylpyrophosphate synthetase-2 associated protein 39 (pap39)
34 c1dkrB_
not modelled
78.6
16
PDB header: transferaseChain: B: PDB Molecule: phosphoribosyl pyrophosphate synthetase;PDBTitle: crystal structures of bacillus subtilis phosphoribosylpyrophosphate2 synthetase: molecular basis of allosteric inhibition and activation.
35 c3hgmD_
not modelled
77.6
15
PDB header: signaling proteinChain: D: PDB Molecule: universal stress protein tead;PDBTitle: universal stress protein tead from the trap transporter teaabc of2 halomonas elongata
36 d2hk6a1
not modelled
77.1
10
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase
37 d1w0ma_
not modelled
76.2
18
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
38 c5t3oB_
not modelled
75.6
17
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase;PDBTitle: crystal structure of the phosphorybosylpyrophosphate synthetase ii2 from thermus thermophilus
39 d1iowa1
not modelled
73.7
22
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain
40 d1tq8a_
not modelled
72.3
27
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: Universal stress protein-like
41 c3fg9B_
not modelled
71.8
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of universal stress protein uspa family;PDBTitle: the crystal structure of an universal stress protein uspa2 family protein from lactobacillus plantarum wcfs1
42 d1ozha1
not modelled
71.0
9
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
43 d2ji7a1
not modelled
69.0
14
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
44 c5j6qA_
not modelled
66.4
25
PDB header: cell adhesionChain: A: PDB Molecule: cell wall binding protein cwp8;PDBTitle: cwp8 from clostridium difficile
45 d2iyva1
not modelled
66.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Shikimate kinase (AroK)
46 c3cgxA_
not modelled
65.3
23
PDB header: transferaseChain: A: PDB Molecule: putative nucleotide-diphospho-sugar transferase;PDBTitle: crystal structure of putative nucleotide-diphospho-sugar transferase2 (yp_389115.1) from desulfovibrio desulfuricans g20 at 1.90 a3 resolution
47 d1to6a_
not modelled
65.2
26
Fold: Glycerate kinase ISuperfamily: Glycerate kinase IFamily: Glycerate kinase I
48 d1uana_
not modelled
64.7
30
Fold: LmbE-likeSuperfamily: LmbE-likeFamily: LmbE-like
49 c5gk2A_
not modelled
63.4
11
PDB header: transferaseChain: A: PDB Molecule: ketosynthase stld;PDBTitle: the structure of the h302a mutant of stld
50 c2ixdB_
not modelled
61.6
24
PDB header: hydrolaseChain: B: PDB Molecule: lmbe-related protein;PDBTitle: crystal structure of the putative deacetylase bc1534 from bacillus2 cereus
51 c2vu2D_
not modelled
61.3
23
PDB header: transferaseChain: D: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: biosynthetic thiolase from z. ramigera. complex with s-pantetheine-11-2 pivalate.
52 d2djia1
not modelled
60.8
22
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
53 c4zhtB_
not modelled
60.6
17
PDB header: isomeraseChain: B: PDB Molecule: bifunctional udp-n-acetylglucosamine 2-epimerase/n-PDBTitle: crystal structure of udp-glcnac 2-epimerase
54 c2zbtB_
not modelled
60.5
16
PDB header: lyaseChain: B: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: crystal structure of pyridoxine biosynthesis protein from thermus2 thermophilus hb8
55 c3s29C_
not modelled
60.5
30
PDB header: transferaseChain: C: PDB Molecule: sucrose synthase 1;PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
56 c3vaaC_
not modelled
59.0
15
PDB header: transferaseChain: C: PDB Molecule: shikimate kinase;PDBTitle: 1.7 angstrom resolution crystal structure of shikimate kinase from2 bacteroides thetaiotaomicron
57 d2ihta1
not modelled
58.9
19
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
58 d1zpda1
not modelled
57.7
19
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
59 c2yzrB_
not modelled
57.1
14
PDB header: lyaseChain: B: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: crystal structure of pyridoxine biosynthesis protein from2 methanocaldococcus jannaschii
60 c3rggD_
not modelled
57.0
9
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase, pure protein;PDBTitle: crystal structure of treponema denticola pure bound to air
61 c3cwcB_
not modelled
56.4
26
PDB header: transferaseChain: B: PDB Molecule: putative glycerate kinase 2;PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
62 d1kaga_
not modelled
56.3
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Shikimate kinase (AroK)
63 c2v4wB_
not modelled
55.5
13
PDB header: transferaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa synthase,PDBTitle: crystal structure of human mitochondrial 3-hydroxy-3-2 methylglutaryl-coenzyme a synthase 2 (hmgcs2)
64 c1zuiA_
not modelled
54.4
21
PDB header: transferaseChain: A: PDB Molecule: shikimate kinase;PDBTitle: structural basis for shikimate-binding specificity of helicobacter2 pylori shikimate kinase
65 c2ebdB_
not modelled
54.4
13
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase iii2 from aquifex aeolicus vf5
66 c3efhB_
not modelled
54.4
20
PDB header: transferaseChain: B: PDB Molecule: ribose-phosphate pyrophosphokinase 1;PDBTitle: crystal structure of human phosphoribosyl pyrophosphate2 synthetase 1
67 d1rkba_
not modelled
54.4
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases
68 d1lbqa_
not modelled
54.3
15
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase
69 c2nv2U_
not modelled
53.9
18
PDB header: lyase/transferaseChain: U: PDB Molecule: pyridoxal biosynthesis lyase pdxs;PDBTitle: structure of the plp synthase complex pdx1/2 (yaad/e) from bacillus2 subtilis
70 d1ovma1
not modelled
53.8
8
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
71 c4lzaB_
not modelled
53.4
24
PDB header: transferaseChain: B: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 thermoanaerobacter pseudethanolicus atcc 33223, nysgrc target 029700.
72 c2z1dA_
not modelled
53.2
19
PDB header: metal binding proteinChain: A: PDB Molecule: hydrogenase expression/formation protein hypd;PDBTitle: crystal structure of [nife] hydrogenase maturation protein, hypd from2 thermococcus kodakaraensis
73 d2gx8a1
not modelled
51.8
19
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
74 c4adsF_
not modelled
51.6
14
PDB header: transferase/transferaseChain: F: PDB Molecule: pyridoxine biosynthetic enzyme pdx1 homologue, putative;PDBTitle: crystal structure of plasmodial plp synthase complex
75 c4o9cC_
not modelled
51.5
28
PDB header: transferaseChain: C: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of beta-ketothiolase (phaa) from ralstonia eutropha2 h16
76 c3loqA_
not modelled
51.4
15
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: universal stress protein;PDBTitle: the crystal structure of a universal stress protein from archaeoglobus2 fulgidus dsm 4304
77 c2gx8B_
not modelled
50.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal structure of bacillus cereus protein related to nif3
78 c3jyfB_
not modelled
49.7
20
PDB header: hydrolaseChain: B: PDB Molecule: 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
79 c2iikA_
not modelled
49.7
18
PDB header: transferaseChain: A: PDB Molecule: 3-ketoacyl-coa thiolase, peroxisomal;PDBTitle: crystal structure of human peroxisomal acetyl-coa acyl transferase 12 (acaa1)
80 d1viaa_
not modelled
49.6
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Shikimate kinase (AroK)
81 c5n6yD_
not modelled
49.4
13
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
82 d1hg3a_
not modelled
49.0
17
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
83 c3femB_
not modelled
48.7
15
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
84 d1e6ca_
not modelled
47.9
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Shikimate kinase (AroK)
85 d1zq1a2
not modelled
47.7
30
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase
86 c6bn2A_
not modelled
47.4
27
PDB header: transferaseChain: A: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of acetyl-coa acetyltransferase from elizabethkingia2 anophelis nuhp1
87 c4eunA_
not modelled
47.1
18
PDB header: transferaseChain: A: PDB Molecule: thermoresistant glucokinase;PDBTitle: crystal structure of a sugar kinase (target efi-502144 from janibacter2 sp. htcc2649), unliganded structure
88 c4wysB_
not modelled
46.8
23
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of thiolase from escherichia coli
89 c4dfeB_
not modelled
46.6
19
PDB header: transferaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase iii2 from burkholderia xenovorans
90 c4dd5A_
not modelled
46.5
24
PDB header: transferaseChain: A: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: biosynthetic thiolase (thla1) from clostridium difficile
91 c3fdiA_
not modelled
45.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein from eubacterium2 ventriosum atcc 27560.
92 c1zq1B_
not modelled
45.8
22
PDB header: lyaseChain: B: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit d;PDBTitle: structure of gatde trna-dependent amidotransferase from2 pyrococcus abyssi
93 d1s3ga1
not modelled
44.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases
94 d2gnpa1
not modelled
44.4
8
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like
95 c3s3tD_
not modelled
44.2
14
PDB header: chaperoneChain: D: PDB Molecule: nucleotide-binding protein, universal stress protein uspaPDBTitle: universal stress protein uspa from lactobacillus plantarum
96 d2ocda1
not modelled
43.9
21
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase
97 d1jlja_
not modelled
43.8
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
98 d1zn7a1
not modelled
43.5
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
99 c6hxgE_
not modelled
43.2
13
PDB header: plant proteinChain: E: PDB Molecule: pyridoxal 5'-phosphate synthase-like subunit pdx1.2;PDBTitle: pdx1.2/pdx1.3 complex (intermediate)
100 c2pt5D_
not modelled
43.2
15
PDB header: transferaseChain: D: PDB Molecule: shikimate kinase;PDBTitle: crystal structure of shikimate kinase (aq_2177) from aquifex aeolicus2 vf5
101 c5g3yA_
not modelled
42.9
21
PDB header: transferaseChain: A: PDB Molecule: adenylate kinse;PDBTitle: crystal structure of adenylate kinase ancestor 1 with zn and adp bound
102 c2eu8B_
not modelled
42.7
25
PDB header: transferaseChain: B: PDB Molecule: adenylate kinase;PDBTitle: crystal structure of a thermostable mutant of bacillus2 subtilis adenylate kinase (q199r)
103 d1zfja1
not modelled
42.6
18
Fold: TIM beta/alpha-barrelSuperfamily: Inosine monophosphate dehydrogenase (IMPDH)Family: Inosine monophosphate dehydrogenase (IMPDH)
104 d2cdna1
not modelled
42.4
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases
105 c3hdtB_
not modelled
42.3
26
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative kinase;PDBTitle: crystal structure of putative kinase from clostridium symbiosum atcc2 14940
106 c2h92C_
not modelled
42.2
11
PDB header: transferaseChain: C: PDB Molecule: cytidylate kinase;PDBTitle: crystal structure of staphylococcus aureus cytidine2 monophosphate kinase in complex with cytidine-5'-3 monophosphate
107 c6aqpA_
not modelled
41.8
25
PDB header: transferaseChain: A: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: aspergillus fumigatus cytosolic thiolase: acetylated enzyme in complex2 with coa and potassium ions
108 c4n45B_
not modelled
41.5
23
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of reduced form of thiolase from clostridium2 acetobutylicum
109 c5bz4K_
not modelled
41.4
27
PDB header: transferaseChain: K: PDB Molecule: beta-ketothiolase;PDBTitle: crystal structure of a t1-like thiolase (coa-complex) from2 mycobacterium smegmatis
110 c4q0mA_
not modelled
41.4
22
PDB header: hydrolaseChain: A: PDB Molecule: l-asparaginase;PDBTitle: crystal structure of pyrococcus furiosus l-asparaginase
111 d1g8fa3
not modelled
41.4
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain
112 c4rbnD_
not modelled
41.3
31
PDB header: transferaseChain: D: PDB Molecule: sucrose synthase:glycosyl transferases group 1;PDBTitle: the crystal structure of nitrosomonas europaea sucrose synthase:2 insights into the evolutionary origin of sucrose metabolism in3 prokaryotes
113 c4pzlC_
not modelled
40.5
11
PDB header: transferaseChain: C: PDB Molecule: adenylate kinase;PDBTitle: the crystal structure of adenylate kinase from francisella tularensis2 subsp. tularensis schu s4
114 d2fywa1
not modelled
40.4
10
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
115 c3dloC_
not modelled
40.3
18
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: universal stress protein;PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
116 d1nmpa_
not modelled
40.1
19
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
117 c3ss6B_
not modelled
39.8
17
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: crystal structure of the bacillus anthracis acetyl-coa2 acetyltransferase
118 c1ub7A_
not modelled
39.7
21
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier protein] synthase;PDBTitle: the crystal analysis of beta-keroacyl-[acyl carrier protein] synthase2 iii (fabh)from thermus thermophilus.
119 c2ar7A_
not modelled
39.7
32
PDB header: transferaseChain: A: PDB Molecule: adenylate kinase 4;PDBTitle: crystal structure of human adenylate kinase 4, ak4
120 c3be4A_
not modelled
39.5
21
PDB header: transferaseChain: A: PDB Molecule: adenylate kinase;PDBTitle: crystal structure of cryptosporidium parvum adenylate kinase cgd5_3360