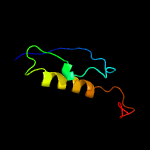
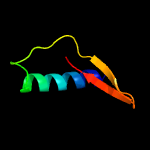
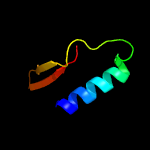
1 c2hhiA_
100.0
48
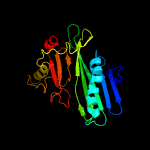
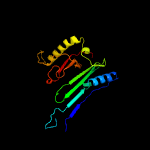
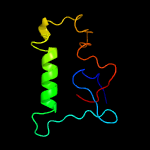
PDB header: unknown functionChain: A: PDB Molecule: immunogenic protein mpt64;PDBTitle: the solution structure of antigen mpt64 from mycobacterium2 tuberculosis defines a novel class of beta-grasp proteins
2 c3cygA_
100.0
18
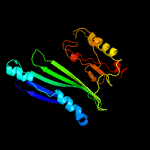
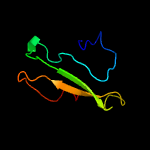
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 fervidobacterium nodosum rt17-b1
3 c5jenA_
100.0
14
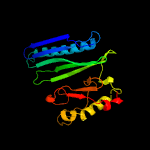
PDB header: hydrolase/hydrolase receptorChain: A: PDB Molecule: anti-sigma-v factor rsiv;PDBTitle: crystal structure of the anti-sigma factor rsiv bound to lysozyme
4 c4e72A_
100.0
13
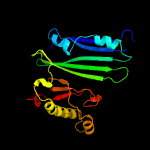
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf3298 family protein (pa4972) from2 pseudomonas aeruginosa pao1 at 2.15 a resolution
5 c3s5tA_
100.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: duf3298 family protein;PDBTitle: crystal structure of a member of duf3298 family (bf2082) from2 bacteroides fragilis nctc 9343 at 2.30 a resolution
6 c4qtqA_
60.6
10
PDB header: hydrolase inhibitorChain: A: PDB Molecule: xac2610 protein;PDBTitle: structure of a xanthomonas type iv secretion system related protein
7 d2c1ia2
44.2
20
Fold: Peptidoglycan deacetylase N-terminal noncatalytic regionSuperfamily: Peptidoglycan deacetylase N-terminal noncatalytic regionFamily: Peptidoglycan deacetylase N-terminal noncatalytic region
8 c3acgA_
29.7
18
PDB header: hydrolaseChain: A: PDB Molecule: beta-1,4-endoglucanase;PDBTitle: crystal structure of carbohydrate-binding module family 28 from2 clostridium josui cel5a in complex with cellobiose
9 c4ot1A_
26.6
12
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: structural basis for the recognition of human cytomegalovirus2 glycoprotein b by the neutralizing human antibody sm5-1
10 d2g5gx1
25.2
12
Fold: EreA/ChaN-likeSuperfamily: EreA/ChaN-likeFamily: ChaN-like
11 d1hf2a2
20.3
18
Fold: Cell-division inhibitor MinC, N-terminal domainSuperfamily: Cell-division inhibitor MinC, N-terminal domainFamily: Cell-division inhibitor MinC, N-terminal domain
12 c2lnzA_
19.7
31
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin-like protein mdy2;PDBTitle: solution structure of the get5 carboxyl domain from s. cerevisiae
13 c3e79A_
19.6
18
PDB header: tpp binding proteinChain: A: PDB Molecule: high affinity transport system protein p37;PDBTitle: structure determination of the cancer-associated mycoplasma2 hyorhinis protein mh-p37
14 c6mjpC_
17.4
6
PDB header: lipid transportChain: C: PDB Molecule: lipopolysaccharide export system protein lptc;PDBTitle: lptb(e163q)fgc from vibrio cholerae
15 d1uwwa_
16.0
5
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 28 carbohydrate binding module, CBM28
16 d2guma1
11.1
2
Fold: Viral glycoprotein ectodomain-likeSuperfamily: Viral glycoprotein ectodomain-likeFamily: Glycoprotein B-like
17 c3vejB_
10.9
29
PDB header: protein bindingChain: B: PDB Molecule: ubiquitin-like protein mdy2;PDBTitle: crystal structure of the get5 carboxyl domain from s. cerevisiae
18 c6ax5A_
10.9
14
PDB header: nuclear proteinChain: A: PDB Molecule: swi/snf-related matrix-associated actin-dependent regulatorPDBTitle: rpt1 region of ini1/snf5/smarcb1_human - swi/snf-related matrix-2 associated actin-dependent regulator of chromatin subfamily b member3 1.
19 c6bm8A_
10.5
2
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
20 c3nw8B_
10.5
2
PDB header: viral proteinChain: B: PDB Molecule: envelope glycoprotein b;PDBTitle: glycoprotein b from herpes simplex virus type 1, y179s mutant, high-ph
21 c2amjD_
not modelled
9.4
10
PDB header: oxidoreductaseChain: D: PDB Molecule: modulator of drug activity b;PDBTitle: crystal structure of modulator of drug activity b from escherichia2 coli o157:h7
22 c5v2sA_
not modelled
8.4
2
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
23 c2hp7A_
not modelled
8.2
21
PDB header: signaling proteinChain: A: PDB Molecule: flagellar motor switch protein flim;PDBTitle: structure of flim provides insight into assembly of the2 switch complex in the bacterial flagella motor
24 d1w6ga3
not modelled
7.9
14
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
25 c3waiA_
not modelled
7.7
21
PDB header: transferase, transport proteinChain: A: PDB Molecule: maltose-binding periplasmic protein, transmembranePDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-l, o29867_arcfu) from archaeoglobus3 fulgidus as a mbp fusion
26 c5c6tA_
not modelled
7.6
9
PDB header: viral protein/immue systemChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of hcmv glycoprotein b in complex with 1g2 fab
27 c2c1iA_
not modelled
7.5
13
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan glcnac deacetylase;PDBTitle: structure of streptococcus pneumoniae peptidoglycan deacetylase2 (sppgda) d 275 n mutant.
28 c4q28B_
not modelled
7.4
7
PDB header: structural proteinChain: B: PDB Molecule: periplakin;PDBTitle: crystal structure of the plectin 1 and 2 repeats of the human2 periplakin. northeast structural genomics consortium (nesg) target3 hr9083a
29 c5yi8B_
not modelled
7.3
33
PDB header: cell cycleChain: B: PDB Molecule: pon peptide from partner of numb;PDBTitle: crystal structure of drosophila numb ptb domain and pon peptide2 complex
30 d3c9fa1
not modelled
7.1
19
Fold: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domainSuperfamily: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domainFamily: 5'-nucleotidase (syn. UDP-sugar hydrolase), C-terminal domain
31 c4yk8A_
not modelled
6.7
19
PDB header: protein transportChain: A: PDB Molecule: meiotically up-regulated gene 66 protein;PDBTitle: crystal structure of the atg101-atg13 complex from fission yeast
32 d1w2za3
not modelled
6.3
14
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
33 c3fvcA_
not modelled
6.3
7
PDB header: viral proteinChain: A: PDB Molecule: glycoprotein gp110;PDBTitle: crystal structure of a trimeric variant of the epstein-barr virus2 glycoprotein b
34 d1to0a_
not modelled
6.2
13
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like
35 d2nysa1
not modelled
5.9
19
Fold: SspB-likeSuperfamily: SspB-likeFamily: AGR C 3712p-like
36 c2nysA_
not modelled
5.9
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: agr_c_3712p;PDBTitle: x-ray crystal structure of protein agr_c_3712 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr88.
37 d1poia_
not modelled
5.4
47
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: CoA transferase alpha subunit-like
38 c6escA_
not modelled
5.3
11
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of pseudorabies virus glycoprotein b
39 c4bk8A_
not modelled
5.3
10
PDB header: oxidoreductaseChain: A: PDB Molecule: desulfoferrodoxin, ferrous iron-binding region;PDBTitle: superoxide reductase (neelaredoxin) from ignicoccus2 hospitalis
40 c3wajA_
not modelled
5.1
22
PDB header: transferaseChain: A: PDB Molecule: transmembrane oligosaccharyl transferase;PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
41 d1u7ba1
not modelled
5.0
10
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor