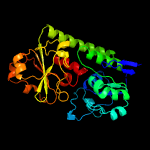
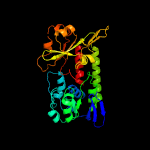
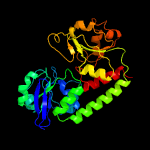
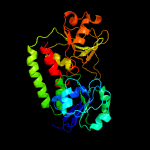
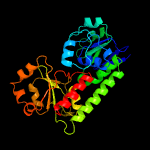
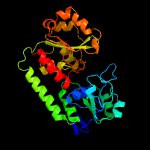
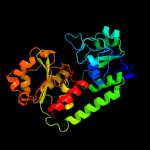
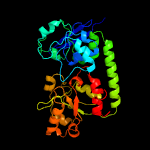
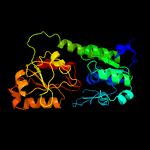
1 c3eiwA_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: htsa protein;PDBTitle: crystal structure of staphylococcus aureus lipoprotein, htsa
2 c3tnyA_
100.0
24
PDB header: metal transportChain: A: PDB Molecule: yfiy (abc transport system substrate-binding protein);PDBTitle: structure of yfiy from bacillus cereus bound to the siderophore iron2 (iii) schizokinen
3 c4hn9B_
100.0
14
PDB header: transport proteinChain: B: PDB Molecule: iron complex transport system substrate-binding protein;PDBTitle: crystal structure of iron abc transporter solute-binding protein from2 eubacterium eligens
4 c3gfvA_
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized abc transporter solute-binding proteinPDBTitle: crystal structure of petrobactin-binding protein yclq from bacillu2 subtilis
5 c3pshA_
100.0
13
PDB header: metal transportChain: A: PDB Molecule: protein hi_1472;PDBTitle: classification of a haemophilus influenzae abc transporter hi1470/712 through its cognate molybdate periplasmic binding protein mola (mola3 bound to molybdate)
6 c5az3A_
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: abc-type transporter, periplasmic component;PDBTitle: crystal structure of heme binding protein hmut
7 c4ovkA_
100.0
16
PDB header: solute-binding proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of periplasmic solute binding protein from2 veillonella parvula dsm 2008
8 c3tefA_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: iron(iii) abc transporter, periplasmic iron-compound-PDBTitle: crystal structure of the periplasmic catecholate-siderophore binding2 protein vctp from vibrio cholerae
9 d2chua1
100.0
20
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TM0189-like
10 c3r5tA_
100.0
19
PDB header: metal transportChain: A: PDB Molecule: ferric vibriobactin abc transporter, periplasmic ferricPDBTitle: crystal structure of holo-viup
11 c3tlkB_
100.0
17
PDB header: metal transportChain: B: PDB Molecule: ferrienterobactin-binding periplasmic protein;PDBTitle: crystal structure of holo fepb
12 c3be5D_
100.0
20
PDB header: metal transportChain: D: PDB Molecule: putative iron compound-binding protein of abc transporterPDBTitle: crystal structure of fite (crystal form 1), a group iii periplasmic2 siderophore binding protein
13 c4jccA_
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: iron-compound abc transporter, iron-compound-bindingPDBTitle: crystal structure of iron uptake abc transporter substrate-binding2 protein piua from streptococcus pneumoniae canada mdr_19a
14 c5cr9A_
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: abc-type fe3+-hydroxamate transport system, periplasmicPDBTitle: crystal structure of abc-type fe3+-hydroxamate transport system from2 saccharomonospora viridis dsm 43017
15 c6mflA_
100.0
19
PDB header: metal transportChain: A: PDB Molecule: baub;PDBTitle: structure of siderophore binding protein baub bound to a complex2 between two molecules of acinetobactin and ferric iron.
16 c6allB_
100.0
18
PDB header: metal transportChain: B: PDB Molecule: fe(3+)-citrate-binding protein yfmc;PDBTitle: crystal structure of a predicted ferric/iron (iii) hydroxymate2 siderophore substrate binding protein from bacillus anthracis
17 c2phzA_
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: iron-uptake system-binding protein;PDBTitle: crystal structure of iron-uptake system-binding protein feua from2 bacillus subtilis. northeast structural genomics target sr580.
18 d2phza1
100.0
14
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TM0189-like
19 c4n01B_
100.0
12
PDB header: transport proteinChain: B: PDB Molecule: periplasmic binding protein;PDBTitle: the crystal structure of a periplasmic binding protein from2 veillonella parvula dsm 2008
20 c4pagA_
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: abc transporter solute binding protein from sulfurospirillum2 deleyianum dsm 6946
21 c5gj3A_
not modelled
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: periplasmic heme-binding protein rhut from roseiflexus sp. rs-1 in2 two-heme bound form (holo-2)
22 c6b2xB_
not modelled
100.0
15
PDB header: metal transportChain: B: PDB Molecule: solute-binding periplasmic protein of iron/siderophore abcPDBTitle: apo yiua crystal form 1
23 c4mx8C_
not modelled
100.0
23
PDB header: solute-binding proteinChain: C: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of troa-like periplasmic binding protein2 peripla_bp_2 from xylanimonas cellulosilytica
24 c4mlzB_
not modelled
100.0
18
PDB header: solute binding proteinChain: B: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of periplasmic binding protein from jonesia2 denitrificans
25 c4ljsA_
not modelled
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: the crystal structure of a periplasmic binding protein from2 veillonella parvula dsm 2008
26 c5dh0B_
not modelled
100.0
20
PDB header: protein bindingChain: B: PDB Molecule: siderophore periplasmic binding protein;PDBTitle: structure of the siderophore periplasmic binding protein from the2 fuscachelin gene cluster of thermobifida fusca in p41
27 c5flyB_
not modelled
100.0
19
PDB header: metal transportChain: B: PDB Molecule: ferrichrome-binding periplasmic protein;PDBTitle: the fhud protein from s.pseudintermedius
28 c4b8yA_
not modelled
100.0
18
PDB header: transport protein/siderophoreChain: A: PDB Molecule: fhud2;PDBTitle: ferrichrome-bound fhud2
29 c4pm4B_
not modelled
100.0
16
PDB header: solute-binding proteinChain: B: PDB Molecule: iron complex transporter substrate-binding protein;PDBTitle: structure of a putative periplasmic iron siderophore binding protein2 (rv0265c) from mycobacterium tuberculosis h37rv
30 c4h59A_
not modelled
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: iron-compound abc transporter, iron compound-bindingPDBTitle: crystal structure of iron uptake abc transporter substrate-binding2 protein piaa from streptococcus pneumoniae canada mdr_19a bound to3 bis-tris propane
31 c4mo9A_
not modelled
100.0
15
PDB header: solute-binding proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of troa-like periplasmic binding protein fepb from2 veillonella parvula
32 c5khlB_
not modelled
100.0
16
PDB header: transport proteinChain: B: PDB Molecule: hemin abc transporter, periplasmic hemin-binding proteinPDBTitle: crystal structure of periplasmic heme binding protein hutb of vibrio2 cholerae
33 c2r79A_
not modelled
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of a periplasmic heme binding protein from2 pseudomonas aeruginosa
34 c5joqA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: lmo2184 protein;PDBTitle: crystal structure of an abc transporter substrate-binding protein from2 listeria monocytogenes egd-e
35 c4mdyA_
not modelled
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of periplasmic solute binding protein from2 mycobacterium smegmatis str. mc2 155
36 c5yscA_
not modelled
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: vitamin b12-binding protein;PDBTitle: crystal structure of periplasmic vitamin b12 binding protein btuf of2 vibrio cholerae
37 c4inpA_
not modelled
100.0
10
PDB header: transport proteinChain: A: PDB Molecule: iron (iii) abc transporter, periplasmic iron-bindingPDBTitle: the crystal structure of helicobacter pylori ceue (hp1561) with ni(ii)2 bound
38 c4m7oA_
not modelled
100.0
14
PDB header: iron binding proteinChain: A: PDB Molecule: iron-binding protein;PDBTitle: the crystal structure of a possible an iron-binding (periplasmic2 solute-binding) protein from staphylococcus epidermidis atcc 12228.
39 c3mwgA_
not modelled
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: iron-regulated abc transporter siderophore-binding proteinPDBTitle: crystal structure of staphylococcus aureus sira
40 c3md9A_
not modelled
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: hemin-binding periplasmic protein hmut;PDBTitle: structure of apo form of a periplasmic heme binding protein
41 c2x4lA_
not modelled
100.0
13
PDB header: transportChain: A: PDB Molecule: ferric-siderophore receptor protein;PDBTitle: crystal structure of dese, a ferric-siderophore receptor protein from2 streptomyces coelicolor
42 c2r7aC_
not modelled
100.0
19
PDB header: transport proteinChain: C: PDB Molecule: bacterial heme binding protein;PDBTitle: crystal structure of a periplasmic heme binding protein from shigella2 dysenteriae
43 c5b58T_
not modelled
100.0
22
PDB header: metal binding proteinChain: T: PDB Molecule: putative hemin transport system, substrate-binding protein;PDBTitle: inward-facing conformation of abc heme importer bhuuv in complex with2 periplasmic heme binding protein bhut from burkholderia cenocepacia
44 c2q8pA_
not modelled
100.0
13
PDB header: metal transportChain: A: PDB Molecule: iron-regulated surface determinant e;PDBTitle: crystal structure of selenomethionine labelled s. aureus isde2 complexed with heme
45 d1esza_
not modelled
100.0
19
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Periplasmic ferric siderophore binding protein FhuD
46 c5ggxD_
not modelled
100.0
21
PDB header: transport proteinChain: D: PDB Molecule: iron(iii) abc transporter, periplasmic iron-compound-PDBTitle: crystal structure of fe3+ - desferal bound siderophore binding protein2 fhud from vibrio cholerae
47 c3g9qA_
not modelled
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: ferrichrome-binding protein;PDBTitle: crystal structure of the fhud fold-family bsu3320, a periplasmic2 binding protein component of a fep/fec-like ferrichrome abc3 transporter from bacillus subtilis. northeast structural genomics4 consortium target sr577a
48 d2etva1
not modelled
100.0
11
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TM0189-like
49 d1n2za_
not modelled
100.0
15
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: TroA-like
50 c5n6yD_
not modelled
93.7
11
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
51 c3aerC_
not modelled
92.7
12
PDB header: oxidoreductaseChain: C: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
52 c2ynmC_
not modelled
92.7
11
PDB header: oxidoreductaseChain: C: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
53 d1m1na_
not modelled
90.9
9
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
54 d1qh8a_
not modelled
89.4
14
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
55 c3pdiG_
not modelled
82.2
13
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
56 d1qh8b_
not modelled
77.9
16
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
57 c2ynmD_
not modelled
72.2
17
PDB header: oxidoreductaseChain: D: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
58 d1m1nb_
not modelled
66.6
15
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
59 c4rvgA_
not modelled
60.5
14
PDB header: transferaseChain: A: PDB Molecule: d-mycarose 3-c-methyltransferase;PDBTitle: crystal structure of mtmc in complex with sam and tdp
60 c5kojD_
not modelled
58.4
18
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase femo beta subunit protein nifk;PDBTitle: nitrogenase mofep protein in the ids oxidized state
61 d1miob_
not modelled
57.7
13
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
62 c3aerB_
not modelled
56.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
63 d1mioa_
not modelled
53.1
6
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
64 c5jm0A_
not modelled
47.3
9
PDB header: hydrolaseChain: A: PDB Molecule: alpha-mannosidase,alpha-mannosidase,alpha-mannosidase;PDBTitle: structure of the s. cerevisiae alpha-mannosidase 1
65 c5vytD_
not modelled
47.1
15
PDB header: transferaseChain: D: PDB Molecule: gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-PDBTitle: crystal structure of the wbkc n-formyltransferase (f142a variant) from2 brucella melitensis
66 c3ndjA_
not modelled
45.4
15
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: x-ray structure of a c-3'-methyltransferase in complex with s-2 adenosyl-l-homocysteine and sugar product
67 d1q3qa2
not modelled
43.3
16
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
68 c3pdiB_
not modelled
36.4
10
PDB header: protein bindingChain: B: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nifn;PDBTitle: precursor bound nifen
69 c2r8rB_
not modelled
35.9
28
PDB header: transferaseChain: B: PDB Molecule: sensor protein;PDBTitle: crystal structure of the n-terminal region (19..243) of sensor protein2 kdpd from pseudomonas syringae pv. tomato str. dc3000
70 d1gmla_
not modelled
35.8
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
71 c3woaA_
not modelled
35.4
13
PDB header: dna binding protein, sugar binding proteChain: A: PDB Molecule: repressor protein ci, maltose-binding periplasmic protein;PDBTitle: crystal structure of lambda repressor (1-45) fused with maltose-2 binding protein
72 d1assa_
not modelled
31.5
16
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
73 d1a6db2
not modelled
30.1
10
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: Group II chaperonin (CCT, TRIC), apical domain
74 d7reqa2
not modelled
30.0
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
75 c2xdqA_
not modelled
26.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
76 d1edza2
not modelled
22.0
7
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase
77 c5ghrA_
not modelled
21.9
15
PDB header: dna binding protein/replicationChain: A: PDB Molecule: ssdna-specific exonuclease;PDBTitle: dna replication protein
78 d1a9xb2
not modelled
21.8
26
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)
79 d1ccwa_
not modelled
20.2
16
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
80 d1a4ia2
not modelled
18.4
9
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Tetrahydrofolate dehydrogenase/cyclohydrolase
81 c4hh3C_
not modelled
18.0
12
PDB header: flavoprotein/transcriptionChain: C: PDB Molecule: appa protein;PDBTitle: structure of the appa-ppsr2 core complex from rb. sphaeroides
82 c5ghrC_
not modelled
17.6
16
PDB header: dna binding protein/replicationChain: C: PDB Molecule: ssdna-specific exonuclease;PDBTitle: dna replication protein
83 c6aqeB_
not modelled
15.8
17
PDB header: transferaseChain: B: PDB Molecule: molecule a;PDBTitle: crystal structure of ppk2 in complex with mg atp
84 c5kzoA_
not modelled
15.6
44
PDB header: transcriptionChain: A: PDB Molecule: neurogenic locus notch homolog protein 1;PDBTitle: notch1 transmembrane and associated juxtamembrane segment
85 c1a4iB_
not modelled
15.5
9
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase /PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
86 c6dspB_
not modelled
14.7
12
PDB header: signaling proteinChain: B: PDB Molecule: autoinducer 2-binding protein lsrb;PDBTitle: lsrb from clostridium saccharobutylicum in complex with ai-2
87 c5n6yE_
not modelled
14.3
9
PDB header: oxidoreductaseChain: E: PDB Molecule: nitrogenase vanadium-iron protein beta chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
88 d1q7ea_
not modelled
13.7
11
Fold: CoA-transferase family III (CaiB/BaiF)Superfamily: CoA-transferase family III (CaiB/BaiF)Family: CoA-transferase family III (CaiB/BaiF)
89 c5o4jC_
not modelled
13.5
19
PDB header: transferaseChain: C: PDB Molecule: hcgc;PDBTitle: hcgc from methanococcus maripaludis cocrystallized with sah and2 pyridinol
90 c2gm2A_
not modelled
13.1
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
91 c4yegA_
not modelled
12.8
6
PDB header: transferaseChain: A: PDB Molecule: polyphosphate kinase 2;PDBTitle: characterisation of polyphosphate kinase 2 from the intracellular2 pathogen francisella tularensis
92 c3czqA_
not modelled
12.8
8
PDB header: transferaseChain: A: PDB Molecule: putative polyphosphate kinase 2;PDBTitle: crystal structure of putative polyphosphate kinase 2 from2 sinorhizobium meliloti
93 c4oxqB_
not modelled
11.4
10
PDB header: metal binding proteinChain: B: PDB Molecule: manganese abc transporter, periplasmic-binding proteinPDBTitle: structure of staphylococcus pseudintermedius metal-binding protein2 sita in complex with zinc
94 c4u7pA_
not modelled
11.3
20
PDB header: transferase/transferase regulatorChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: crystal structure of dnmt3a-dnmt3l complex
95 c4k40B_
not modelled
11.1
31
PDB header: hydrolaseChain: B: PDB Molecule: gdsl-like lipase/acylhydrolase family protein;PDBTitle: peptidoglycan o-acetylesterase in action, 0 min
96 c5d5oE_
not modelled
11.0
34
PDB header: unknown functionChain: E: PDB Molecule: uncharacterized protein mj0489;PDBTitle: hcgc from methanocaldococcus jannaschii
97 c2z4dA_
not modelled
10.9
20
PDB header: nuclear proteinChain: A: PDB Molecule: 26s proteasome regulatory subunit rpn13;PDBTitle: nmr structures of yeast proteasome component rpn13
98 c3dcjA_
not modelled
10.8
24
PDB header: transferaseChain: A: PDB Molecule: probable 5'-phosphoribosylglycinamide formyltransferasePDBTitle: crystal structure of glycinamide formyltransferase (purn) from2 mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-3 tetrahydrofolic acid derivative
99 c3obiC_
not modelled
10.4
19
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution