1 c2lkyA_
100.0
38
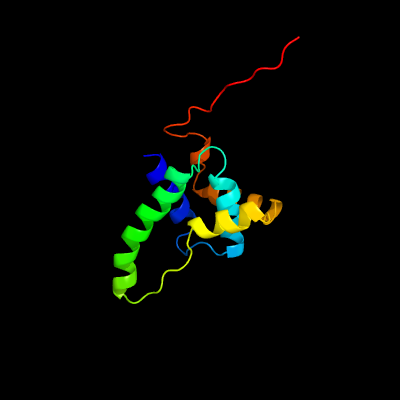
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
2 c2kvcA_
100.0
38
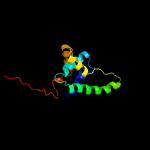
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
3 c3ol4B_
100.0
37
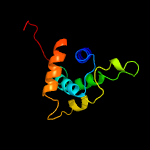
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
4 c2m0nA_
100.0
54
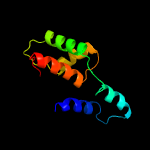
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of a duf3349 annotated protein from mycobacterium2 abscessus, mab_3403c. seattle structural genomics center for3 infectious disease target myaba.17112.a.a2
5 d1khda1
86.2
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
6 d1o17a1
86.1
9
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
7 d1hj3a1
83.0
30
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
8 d1v8ga1
79.4
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
9 d1qksa1
79.3
21
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
10 d1uoua1
76.9
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
11 d2tpta1
73.0
18
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
12 d1brwa1
71.8
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
13 c3mk7F_
61.6
9
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
14 c1o17A_
61.2
8
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
15 d1e2rb1
58.7
16
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
16 c4hkmA_
57.2
6
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: crystal structure of an anthranilate phosphoribosyltransferase (target2 id nysgrc-016600) from xanthomonas campestris
17 c1ta9A_
57.1
19
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol dehydrogenase;PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
18 d1nira1
54.0
26
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
19 d1t1ea2
51.6
21
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors
20 c3ee6A_
50.3
21
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure analysis of tripeptidyl peptidase -i
21 d2c8sa1
not modelled
50.1
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
22 c2m29A_
not modelled
49.0
16
PDB header: metal binding proteinChain: A: PDB Molecule: calcium-binding protein 4;PDBTitle: nmr structure of ca2+ bound cabp4 n-domain
23 c3ek7A_
not modelled
47.6
26
PDB header: fluorescent proteinChain: A: PDB Molecule: myosin light chain kinase, green fluorescent protein,PDBTitle: calcium-saturated gcamp2 dimer
24 c3gr0D_
not modelled
46.9
18
PDB header: membrane proteinChain: D: PDB Molecule: protein prgh;PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
25 d1avsa_
not modelled
46.2
18
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
26 c3edyA_
not modelled
44.9
22
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure of the precursor form of human tripeptidyl-peptidase2 1
27 c4oy4A_
not modelled
44.6
23
PDB header: calcium binding, fluorescent, transferasChain: A: PDB Molecule: chimera protein of calmodulin, gpf-like protein eosfp, andPDBTitle: calcium-free campari v0.2
28 d1ng6a_
not modelled
44.4
15
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/YqeY domain
29 c2k7bA_
not modelled
43.9
12
PDB header: metal binding proteinChain: A: PDB Molecule: calcium-binding protein 1;PDBTitle: nmr structure of mg2+-bound cabp1 n-domain
30 c1khdD_
not modelled
42.5
17
PDB header: transferaseChain: D: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.9 resolution3 (current name, pectobacterium carotovorum)
31 c4muoB_
not modelled
41.1
8
PDB header: dna binding proteinChain: B: PDB Molecule: uncharacterized protein ybib;PDBTitle: the trpd2 enzyme from e.coli: ybib
32 d1h9xa1
not modelled
40.9
18
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
33 d2pq3a1
not modelled
40.3
25
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
34 c2ktgA_
not modelled
40.2
13
PDB header: ca-binding proteinChain: A: PDB Molecule: calmodulin, putative;PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein
35 c1t1eA_
not modelled
39.9
18
PDB header: hydrolaseChain: A: PDB Molecule: kumamolisin;PDBTitle: high resolution crystal structure of the intact pro-2 kumamolisin, a sedolisin type proteinase (previously3 called kumamolysin or kscp)
36 c6ge2A_
not modelled
39.4
26
PDB header: hormoneChain: A: PDB Molecule: exendin-4;PDBTitle: exendin-4 based dual glp-1/glucagon receptor agonist
37 c2zxyA_
not modelled
38.8
12
PDB header: oxygen binding, transport proteinChain: A: PDB Molecule: cytochrome c552;PDBTitle: crystal structure of cytochrome c555 from aquifex aeolicus
38 c3u0kA_
not modelled
37.8
18
PDB header: fluorescent proteinChain: A: PDB Molecule: rcamp;PDBTitle: crystal structure of the genetically encoded calcium indicator rcamp
39 c1brwB_
not modelled
37.4
20
PDB header: transferaseChain: B: PDB Molecule: protein (pyrimidine nucleoside phosphorylase);PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
40 d1hzua1
not modelled
36.7
26
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase
41 c2dsjA_
not modelled
36.2
10
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside (thymidine) phosphorylase;PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
42 c1jrjA_
not modelled
35.9
42
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
43 c2xtsD_
not modelled
33.8
14
PDB header: oxidoreductase/electron transportChain: D: PDB Molecule: cytochrome;PDBTitle: crystal structure of the sulfane dehydrogenase soxcd from paracoccus2 pantotrophus
44 c5n1tB_
not modelled
33.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c;PDBTitle: crystal structure of complex between flavocytochrome c and copper2 chaperone copc from t. paradoxus
45 c3cuqD_
not modelled
33.2
27
PDB header: protein transportChain: D: PDB Molecule: vacuolar protein-sorting-associated protein 25;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
46 c2zmeD_
not modelled
33.2
27
PDB header: protein transportChain: D: PDB Molecule: vacuolar protein-sorting-associated protein 25;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
47 c5mwwA_
not modelled
32.4
25
PDB header: transferaseChain: A: PDB Molecule: rna polymerase sigma factor siga;PDBTitle: sigma1.1 domain of sigmaa from bacillus subtilis
48 d1dvha_
not modelled
31.5
9
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
49 c1h1oA_
not modelled
30.5
14
PDB header: electron transportChain: A: PDB Molecule: cytochrome c-552;PDBTitle: acidithiobacillus ferrooxidans cytochrome c4 structure2 supports a complex-induced tuning of electron transfer
50 c1wvoA_
not modelled
30.4
41
PDB header: transferaseChain: A: PDB Molecule: sialic acid synthase;PDBTitle: solution structure of rsgi ruh-029, an antifreeze protein2 like domain in human n-acetylneuraminic acid phosphate3 synthase gene.
51 c4n6cB_
not modelled
29.6
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the b1rzq2 protein from streptococcus pneumoniae.2 northeast structural genomics consortium (nesg) target spr36.
52 c5zorA_
not modelled
29.3
29
PDB header: metal binding proteinChain: A: PDB Molecule: centrin, putative;PDBTitle: solution structure of centrin4 from trypanosoma brucei
53 c6jtdB_
not modelled
29.2
12
PDB header: transferaseChain: B: PDB Molecule: c-glycosyltransferase;PDBTitle: crystal structure of tccgt1 in complex with udp
54 c4ga5H_
not modelled
27.4
18
PDB header: transferaseChain: H: PDB Molecule: putative thymidine phosphorylase;PDBTitle: crystal structure of amp phosphorylase c-terminal deletion mutant in2 the apo-form
55 c1c8aA_
not modelled
27.3
22
PDB header: antifreeze proteinChain: A: PDB Molecule: protein (antifreeze protein type iii);PDBTitle: nmr structure of intramolecular dimer antifreeze protein2 rd3, 40 sa structures
56 c3h5qA_
not modelled
26.9
13
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside phosphorylase;PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
57 c4i66A_
not modelled
26.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein hoch_4089;PDBTitle: crystal structure of hoch_4089 protein from haliangium ochraceum
58 d3nlaa_
not modelled
26.4
28
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
59 d1c8aa2
not modelled
26.2
22
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
60 d1xb4a1
not modelled
25.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain
61 c5tdyD_
not modelled
25.0
23
PDB header: motor proteinChain: D: PDB Molecule: flagellar motor switch protein flig;PDBTitle: structure of cofolded flifc:flign complex from thermotoga maritima
62 c5xj0Y_
not modelled
24.9
33
PDB header: transferase/transcriptionChain: Y: PDB Molecule: gp76;PDBTitle: t. thermophilus rna polymerase holoenzyme bound with gp39 and gp76
63 d2vcha1
not modelled
24.8
7
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like
64 c2k6xA_
not modelled
24.6
17
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
65 c2bpqB_
not modelled
24.2
16
PDB header: transferaseChain: B: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: anthranilate phosphoribosyltransferase (trpd) from2 mycobacterium tuberculosis (apo structure)
66 c3kblA_
not modelled
24.1
36
PDB header: protein bindingChain: A: PDB Molecule: female germline-specific tumor suppressor gld-1;PDBTitle: crystal structure of the gld-1 homodimerization domain from2 caenorhabditis elegans n169a mutant at 2.28 a resolution
67 d1fcdc1
not modelled
23.9
16
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c
68 c2j0fC_
not modelled
23.9
16
PDB header: transferaseChain: C: PDB Molecule: thymidine phosphorylase;PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
69 c4j20B_
not modelled
23.8
29
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c-555;PDBTitle: x-ray structure of the cytochrome c-554 from chlorobaculum tepidum
70 c1vquB_
not modelled
23.7
12
PDB header: transferaseChain: B: PDB Molecule: anthranilate phosphoribosyltransferase 2;PDBTitle: crystal structure of anthranilate phosphoribosyltransferase 22 (17130499) from nostoc sp. at 1.85 a resolution
71 c3ecjC_
not modelled
23.5
13
PDB header: oxidoreductaseChain: C: PDB Molecule: protein (homoprotocatechuate 2,3-dioxygenase);PDBTitle: structure of e323l mutant of homoprotocatechuate 2,3-dioxygenase from2 brevibacterium fuscum at 1.65a resolution
72 c1zswA_
not modelled
23.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: glyoxalase family protein;PDBTitle: crystal structure of bacillus cereus metallo protein from glyoxalase2 family
73 d1c53a_
not modelled
23.2
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
74 d1c75a_
not modelled
23.2
21
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
75 c4dnnB_
not modelled
22.7
36
PDB header: splicingChain: B: PDB Molecule: protein quaking;PDBTitle: crystal structure of the quaking qua1 homodimerization domain
76 d2acva1
not modelled
22.5
18
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like
77 c3deeA_
not modelled
22.4
8
PDB header: transcriptionChain: A: PDB Molecule: putative regulatory protein;PDBTitle: crystal structure of a putative regulatory protein involved in2 transcription (ngo1945) from neisseria gonorrhoeae fa 1090 at 2.25 a3 resolution
78 c4i2yB_
not modelled
22.3
24
PDB header: fluorescent proteinChain: B: PDB Molecule: rgeco1;PDBTitle: crystal structure of the genetically encoded calcium indicator rgeco1
79 c2zzsW_
not modelled
22.3
14
PDB header: electron transportChain: W: PDB Molecule: PDBTitle: crystal structure of cytochrome c554 from vibrio2 parahaemolyticus strain rimd2210633
80 c2zooA_
not modelled
22.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nitrite reductase;PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
81 d1zaca_
not modelled
22.0
19
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
82 c3omdB_
not modelled
21.8
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
83 c6hwhj_
not modelled
21.8
21
PDB header: electron transportChain: J: PDB Molecule: co-purified unknown peptide built as polyala;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
84 d1opsa_
not modelled
21.4
28
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
85 d1a56a_
not modelled
21.4
9
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
86 c2lv7A_
not modelled
21.4
13
PDB header: metal binding proteinChain: A: PDB Molecule: calcium-binding protein 7;PDBTitle: solution structure of ca2+-bound cabp7 n-terminal doman
87 d1zswa2
not modelled
21.1
9
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: BC1024-like
88 c4ax3A_
not modelled
21.0
6
PDB header: oxidoreductaseChain: A: PDB Molecule: copper-containing nitrite reductase;PDBTitle: structure of three-domain heme-cu nitrite reductase from ralstonia2 pickettii at 1.6 a resolution
89 d1m70a2
not modelled
20.3
11
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c
90 c2ybvN_
not modelled
20.0
28
PDB header: lyaseChain: N: PDB Molecule: ribulose bisphosphate carboxylase small subunit;PDBTitle: structure of rubisco from thermosynechococcus elongatus
91 c6adqC_
not modelled
19.9
18
PDB header: electron transportChain: C: PDB Molecule: cytochrome bc1 complex cytochrome c subunit;PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
92 c2n39A_
not modelled
19.9
27
PDB header: dna binding proteinChain: A: PDB Molecule: chromodomain-helicase-dna-binding protein 1;PDBTitle: nmr solution structure of a c-terminal domain of the chromodomain2 helicase dna-binding protein 1
93 c5v2kA_
not modelled
19.8
12
PDB header: transferaseChain: A: PDB Molecule: udp-glycosyltransferase 74f2;PDBTitle: crystal structure of udp-glucosyltransferase, ugt74f2 (t15a), with udp2 and 2-bromobenzoic acid
94 c4pwaA_
not modelled
19.8
24
PDB header: electron transportChain: A: PDB Molecule: putative cytochrome c;PDBTitle: crystal structure of the c-type cytochrome soru from sinorhizobium2 meliloti
95 d1rbli_
not modelled
19.8
32
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit
96 c2i2rK_
not modelled
19.7
40
PDB header: transport proteinChain: K: PDB Molecule: potassium voltage-gated channel subfamily d member 3;PDBTitle: crystal structure of the kchip1/kv4.3 t1 complex
97 d1f54a_
not modelled
19.5
23
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like
98 d2ji7a1
not modelled
19.2
22
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
99 d2ns0a1
not modelled
19.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like