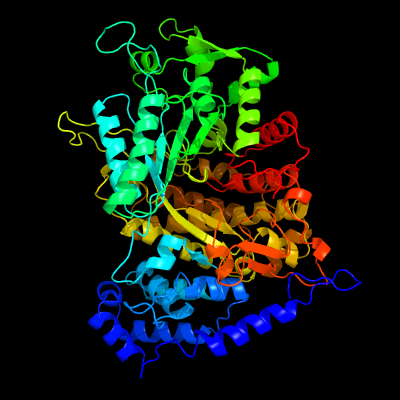
| 1 | c1pemA_
|
|
 |
100.0 |
72 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase 2 alpha
PDBTitle: ribonucleotide reductase protein r1e from salmonella2 typhimurium
|
|
|
|
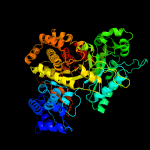
| 2 | c6cgmA_
|
|
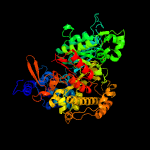 |
100.0 |
48 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase;
PDBTitle: x-ray crystal structure of bacillus subtilis ribonucleotide reductase2 nrde alpha subunit (nucleotide free)
|
|
|
|
| 3 | c3r1rB_
|
|
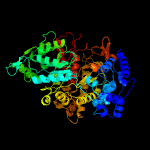 |
100.0 |
24 |
PDB header:complex (oxidoreductase/peptide)
Chain: B: PDB Molecule:ribonucleotide reductase r1 protein;
PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
|
|
|
|
| 4 | c1zyzA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large chain 1;
PDBTitle: structures of yeast ribonucloetide reductase i
|
|
|
|
| 5 | c2wghA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large
PDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
|
|
|
|
| 6 | c2cvuA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large chain
PDBTitle: structures of yeast ribonucleotide reductase i
|
|
|
|
| 7 | c5im3A_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase;
PDBTitle: crystal structure of the class i ribonucleotide reductase from2 pseudomonas aeruginosa in complex with datp
|
|
|
|
| 8 | c3hnfA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large subunit;
PDBTitle: crystal structure of human ribonucleotide reductase 1 bound to the2 effectors ttp and datp
|
|
|
|
| 9 | c3rsrA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase large chain 1;
PDBTitle: crystal structure of 5-nitp inhibition of yeast ribonucleotide2 reductase
|
|
|
|
| 10 | c3pawD_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:ribonucleoside-diphosphate reductase large chain 1;
PDBTitle: low resolution x-ray crystal structure of yeast rnr1p with datp bound2 in the a-site
|
|
|
|
| 11 | c1xjeA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleotide reductase, b12-dependent;
PDBTitle: structural mechanism of allosteric substrate specificity in a2 ribonucleotide reductase: dttp-gdp complex
|
|
|
|
| 12 | d1rlra2
|
|
 |
100.0 |
27 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
|
|
|
| 13 | d1peqa2
|
|
 |
100.0 |
74 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:R1 subunit of ribonucleotide reductase, C-terminal domain |
|
|
|
| 14 | c6dqxA_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside-diphosphate reductase, alpha chain;
PDBTitle: actinobacillus ureae class id ribonucleotide reductase alpha subunit
|
|
|
|
| 15 | c6dqwB_
|
|
 |
100.0 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ribonucleoside-diphosphate reductase, alpha chain;
PDBTitle: flavobacterium johnsoniae class id ribonucleotide reductase alpha2 subuint
|
|
|
|
| 16 | c6dqwD_
|
|
 |
100.0 |
35 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:ribonucleoside-diphosphate reductase, alpha chain;
PDBTitle: flavobacterium johnsoniae class id ribonucleotide reductase alpha2 subuint
|
|
|
|
| 17 | d1l1la_
|
|
 |
100.0 |
19 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:B12-dependent (class II) ribonucleotide reductase |
|
|
|
| 18 | d1peqa1
|
|
 |
100.0 |
64 |
Fold:R1 subunit of ribonucleotide reductase, N-terminal domain
Superfamily:R1 subunit of ribonucleotide reductase, N-terminal domain
Family:R1 subunit of ribonucleotide reductase, N-terminal domain |
|
|
|
| 19 | d1rlra1
|
|
 |
100.0 |
11 |
Fold:R1 subunit of ribonucleotide reductase, N-terminal domain
Superfamily:R1 subunit of ribonucleotide reductase, N-terminal domain
Family:R1 subunit of ribonucleotide reductase, N-terminal domain |
|
|
|
| 20 | c4u3eA_
|
|
 |
98.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside triphosphate reductase;
PDBTitle: anaerobic ribonucleotide reductase
|
|
|
|
| 21 | c1hk8A_ |
|
not modelled |
97.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity regulation in2 class iii ribonucleotide reductases: nrdd in complex with dgtp
|
|
|
| 22 | d1hk8a_ |
|
not modelled |
97.4 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
|
|
| 23 | c1h7bA_ |
|
not modelled |
97.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase large
PDBTitle: structural basis for allosteric substrate specificity regulation in2 class iii ribonucleotide reductases, native nrdd
|
|
|
| 24 | c3pg6D_ |
|
not modelled |
59.7 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:e3 ubiquitin-protein ligase dtx3l;
PDBTitle: the carboxyl terminal domain of human deltex 3-like
|
|
|
| 25 | d1h16a_ |
|
not modelled |
57.7 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
|
|
| 26 | d2okqa1 |
|
not modelled |
51.4 |
22 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YbaA-like |
|
|
| 27 | c3vndD_ |
|
not modelled |
47.1 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 28 | c4ov9A_ |
|
not modelled |
41.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:isopropylmalate synthase;
PDBTitle: structure of isopropylmalate synthase binding with alpha-2 isopropylmalate
|
|
|
| 29 | c2okqB_ |
|
not modelled |
41.1 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ybaa;
PDBTitle: crystal structure of unknown conserved ybaa protein from shigella2 flexneri
|
|
|
| 30 | c5ey5A_ |
|
not modelled |
39.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 31 | d1h3ob_ |
|
not modelled |
36.3 |
9 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
|
|
| 32 | c3p9aI_ |
|
not modelled |
35.9 |
11 |
PDB header:dna binding protein
Chain: I: PDB Molecule:dna-packaging protein gp3;
PDBTitle: an atomic view of the nonameric small terminase subunit of2 bacteriophage p22
|
|
|
| 33 | d2ccqa1 |
|
not modelled |
31.9 |
12 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
|
|
| 34 | c5udbB_ |
|
not modelled |
29.5 |
11 |
PDB header:replication
Chain: B: PDB Molecule:origin recognition complex subunit 2;
PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
|
|
|
| 35 | c4juyB_ |
|
not modelled |
27.4 |
8 |
PDB header:unknown function
Chain: B: PDB Molecule:e3 ubiquitin-protein ligase rnf31;
PDBTitle: crystal structure of the pub domain of e3 ubiquitin ligase rnf31
|
|
|
| 36 | d1ic8a2 |
|
not modelled |
27.1 |
17 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:POU-specific domain |
|
|
| 37 | d1geqa_ |
|
not modelled |
26.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 38 | c6hqaK_ |
|
not modelled |
25.8 |
10 |
PDB header:transcription
Chain: K: PDB Molecule:subunit (61/68 kda) of tfiid and saga complexes;
PDBTitle: molecular structure of promoter-bound yeast tfiid
|
|
|
| 39 | d1qhma_ |
|
not modelled |
25.8 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
|
|
| 40 | c5kzmA_ |
|
not modelled |
24.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 41 | c3u3iA_ |
|
not modelled |
23.9 |
24 |
PDB header:rna binding protein
Chain: A: PDB Molecule:nucleocapsid protein;
PDBTitle: a rna binding protein from crimean-congo hemorrhagic fever virus
|
|
|
| 42 | d1v93a_ |
|
not modelled |
21.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FAD-linked oxidoreductase
Family:Methylenetetrahydrofolate reductase |
|
|
| 43 | c5tchG_ |
|
not modelled |
21.4 |
14 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 44 | c4xgcB_ |
|
not modelled |
18.7 |
15 |
PDB header:dna binding protein
Chain: B: PDB Molecule:origin recognition complex subunit 2;
PDBTitle: crystal structure of the eukaryotic origin recognition complex
|
|
|
| 45 | c6fosO_ |
|
not modelled |
17.5 |
10 |
PDB header:photosynthesis
Chain: O: PDB Molecule:psam;
PDBTitle: cyanidioschyzon merolae photosystem i
|
|
|
| 46 | c5vodD_ |
|
not modelled |
17.3 |
19 |
PDB header:viral protein/immune system
Chain: D: PDB Molecule:envelope glycoprotein ul130;
PDBTitle: crystal structure of hcmv pentamer in complex with neutralizing2 antibody 9i6
|
|
|
| 47 | c3navB_ |
|
not modelled |
15.8 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 48 | c5ze4A_ |
|
not modelled |
15.6 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
|
|
|
| 49 | d1msza_ |
|
not modelled |
15.6 |
14 |
Fold:IF3-like
Superfamily:R3H domain
Family:R3H domain |
|
|
| 50 | c1mszA_ |
|
not modelled |
15.6 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein smubp-2;
PDBTitle: solution structure of the r3h domain from human smubp-2
|
|
|
| 51 | d2d5ua1 |
|
not modelled |
15.1 |
12 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
|
|
| 52 | c2xrgA_ |
|
not modelled |
14.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
|
|
| 53 | d1vmha_ |
|
not modelled |
14.7 |
13 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
|
|
| 54 | d1vmfa_ |
|
not modelled |
14.4 |
6 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
|
|
| 55 | d1vmja_ |
|
not modelled |
14.1 |
23 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
|
|
| 56 | c3kowH_ |
|
not modelled |
13.8 |
23 |
PDB header:metal binding protein
Chain: H: PDB Molecule:d-ornithine aminomutase s component;
PDBTitle: crystal structure of ornithine 4,5 aminomutase backsoaked complex
|
|
|
| 57 | d1td6a_ |
|
not modelled |
13.7 |
23 |
Fold:Hypothetical protein MPN330
Superfamily:Hypothetical protein MPN330
Family:Hypothetical protein MPN330 |
|
|
| 58 | c4pfxA_ |
|
not modelled |
13.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase (galt1);
PDBTitle: the highly conserved domain of unknown function 1792 has a distinct2 glycosyltransferase fold
|
|
|
| 59 | d1vkna2 |
|
not modelled |
13.4 |
14 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
| 60 | c2i3eA_ |
|
not modelled |
13.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:g-rich;
PDBTitle: solution structure of catalytic domain of goldfish rich2 protein
|
|
|
| 61 | c5v4aB_ |
|
not modelled |
12.5 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyltransferase (duf1792);
PDBTitle: a new glycosyltransferase (duf1792) from streptococcus sanguinis
|
|
|
| 62 | d1v74b_ |
|
not modelled |
12.4 |
16 |
Fold:Four-helical up-and-down bundle
Superfamily:Colicin D immunity protein
Family:Colicin D immunity protein |
|
|
| 63 | c3rcqA_ |
|
not modelled |
12.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartyl/asparaginyl beta-hydroxylase;
PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
|
|
|
| 64 | c2p6hB_ |
|
not modelled |
11.8 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
|
|
|
| 65 | c2v1nA_ |
|
not modelled |
11.7 |
19 |
PDB header:nuclear protein
Chain: A: PDB Molecule:protein kin homolog;
PDBTitle: solution structure of the region 51-160 of human kin172 reveals a winged helix fold
|
|
|
| 66 | c2xr9A_ |
|
not modelled |
11.4 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2)
|
|
|
| 67 | c5c8hA_ |
|
not modelled |
11.4 |
30 |
PDB header:replication
Chain: A: PDB Molecule:origin recognition complex subunit 2;
PDBTitle: crystal structure of orc2 c-terminal domain
|
|
|
| 68 | c4b56A_ |
|
not modelled |
11.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: structure of ectonucleotide pyrophosphatase-phosphodiesterase-12 (npp1)
|
|
|
| 69 | c5jqyA_ |
|
not modelled |
11.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartyl/asparaginyl beta-hydroxylase;
PDBTitle: aspartyl/asparaginyl beta-hydroxylase (asph)oxygenase and tpr domains2 in complex with manganese, n-oxalylglycine and factor x substrate3 peptide fragment(39mer-4ser)
|
|
|
| 70 | c5ym0A_ |
|
not modelled |
10.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the crystal structure of dhad
|
|
|
| 71 | c3oiyB_ |
|
not modelled |
10.4 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:reverse gyrase helicase domain;
PDBTitle: helicase domain of reverse gyrase from thermotoga maritima
|
|
|
| 72 | d1vpha_ |
|
not modelled |
10.4 |
11 |
Fold:YjbQ-like
Superfamily:YjbQ-like
Family:YjbQ-like |
|
|
| 73 | c5ujmB_ |
|
not modelled |
10.2 |
30 |
PDB header:replication
Chain: B: PDB Molecule:origin recognition complex subunit 2;
PDBTitle: structure of the active form of human origin recognition complex and2 its atpase motor module
|
|
|
| 74 | c5v89A_ |
|
not modelled |
10.0 |
21 |
PDB header:ligase / protein binding
Chain: A: PDB Molecule:dcn1-like protein 4;
PDBTitle: structure of dcn4 pony domain bound to cul1 whb
|
|
|
| 75 | c3ty3A_ |
|
not modelled |
10.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable homoisocitrate dehydrogenase;
PDBTitle: crystal structure of homoisocitrate dehydrogenase from2 schizosaccharomyces pombe bound to glycyl-glycyl-glycine
|
|
|
| 76 | c3pe0B_ |
|
not modelled |
9.9 |
36 |
PDB header:structural protein
Chain: B: PDB Molecule:plectin;
PDBTitle: structure of the central region of the plakin domain of plectin
|
|
|
| 77 | c5i2gB_ |
|
not modelled |
9.9 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:diol dehydratase;
PDBTitle: 1,2-propanediol dehydration in roseburia inulinivorans; structural2 basis for substrate and enantiomer selectivity
|
|
|
| 78 | c2p6cB_ |
|
not modelled |
9.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:aq_2013 protein;
PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
|
|
|
| 79 | c5j84A_ |
|
not modelled |
9.8 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase;
PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
|
|
|
| 80 | c2l09A_ |
|
not modelled |
9.6 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:asr4154 protein;
PDBTitle: solution nmr structure of protein asr4154 from nostoc sp. pcc71202 northeast structural genomics consortium target id nsr143
|
|
|
| 81 | d2d0ob1 |
|
not modelled |
9.4 |
19 |
Fold:Anticodon-binding domain-like
Superfamily:B12-dependent dehydatase associated subunit
Family:Dehydratase-reactivating factor beta subunit |
|
|
| 82 | c3h1yA_ |
|
not modelled |
9.3 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose 6-phosphate isomerase from2 salmonella typhimurium bound to substrate (f6p)and metal3 atom (zn)
|
|
|
| 83 | c5xgqB_ |
|
not modelled |
9.0 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:methionine-trna ligase;
PDBTitle: crystal structure of apo form (free-state) mycobacterium tuberculosis2 methionyl-trna synthetase
|
|
|
| 84 | c3q7cA_ |
|
not modelled |
8.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleoprotein;
PDBTitle: exonuclease domain of lassa virus nucleoprotein bound to manganese
|
|
|
| 85 | d2b4va2 |
|
not modelled |
8.9 |
33 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:RNA editing terminal uridyl transferase 2, RET2, catalytic domain |
|
|
| 86 | c5wqlC_ |
|
not modelled |
8.8 |
15 |
PDB header:protein binding/signaling protein/hydrol
Chain: C: PDB Molecule:tail-specific protease;
PDBTitle: structure of a pdz-protease bound to a substrate-binding adaptor
|
|
|
| 87 | c4cx8B_ |
|
not modelled |
8.7 |
50 |
PDB header:viral protein
Chain: B: PDB Molecule:pseudorabies virus protease;
PDBTitle: monomeric pseudorabies virus protease pul26n at 2.5 a resolution
|
|
|
| 88 | d1vkia_ |
|
not modelled |
8.6 |
7 |
Fold:YbaK/ProRS associated domain
Superfamily:YbaK/ProRS associated domain
Family:YbaK/ProRS associated domain |
|
|
| 89 | c2c1fA_ |
|
not modelled |
8.5 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:bifunctional endo-1,4-beta-xylanase a;
PDBTitle: the structure of the family 11 xylanase from neocallimastix2 patriciarum
|
|
|
| 90 | c4djzH_ |
|
not modelled |
8.5 |
31 |
PDB header:hydrolase/hydrolase inhibitor
Chain: H: PDB Molecule:protease inhibitor sgpi-2;
PDBTitle: catalytic fragment of masp-1 in complex with its specific inhibitor2 developed by directed evolution on sgci scaffold
|
|
|
| 91 | c5d9aD_ |
|
not modelled |
8.2 |
13 |
PDB header:viral protein
Chain: D: PDB Molecule:polymerase acidic protein;
PDBTitle: influenza c virus rna-dependent rna polymerase - space group p212121
|
|
|
| 92 | c3w53A_ |
|
not modelled |
8.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of psychrophilic beta-glucosidase bglu from2 micrococcus antarcticus
|
|
|
| 93 | d1j09a2 |
|
not modelled |
8.1 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
|
|
| 94 | c5dmuA_ |
|
not modelled |
8.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:nhej polymerase;
PDBTitle: structure of the nhej polymerase from methanocella paludicola
|
|
|
| 95 | d1at3a_ |
|
not modelled |
8.0 |
40 |
Fold:Herpes virus serine proteinase, assemblin
Superfamily:Herpes virus serine proteinase, assemblin
Family:Herpes virus serine proteinase, assemblin |
|
|
| 96 | c3ig3A_ |
|
not modelled |
7.9 |
22 |
PDB header:signaling protein, membrane protein
Chain: A: PDB Molecule:plxna3 protein;
PDBTitle: crystal structure of mouse plexin a3 intracellular domain
|
|
|
| 97 | c1eu1A_ |
|
not modelled |
7.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethyl sulfoxide reductase;
PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
|
|
|
| 98 | d1wiga2 |
|
not modelled |
7.8 |
33 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
|
|
| 99 | c2v3sB_ |
|
not modelled |
7.8 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:serine/threonine-protein kinase osr1;
PDBTitle: structural insights into the recognition of substrates and2 activators by the osr1 kinase
|
|
|