| 1 |
|
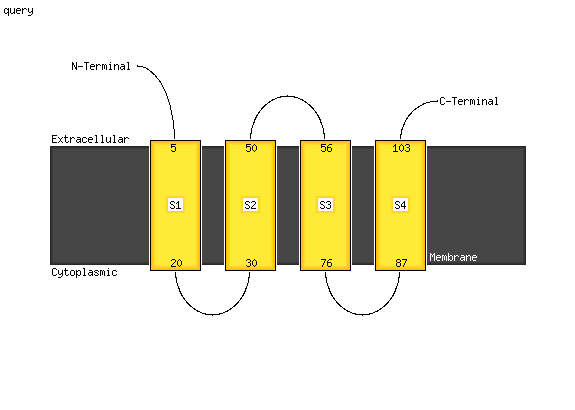
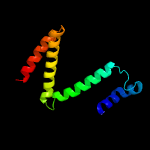
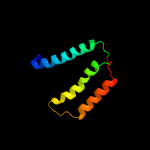
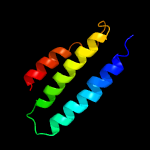
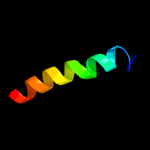
PDB 1s7b chain A
Region: 4 - 105
Aligned: 101
Modelled: 102
Confidence: 100.0%
Identity: 43%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 2 |
|
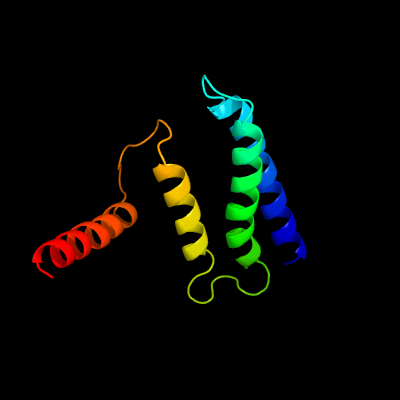
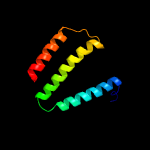
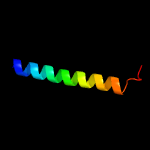
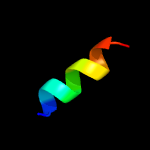
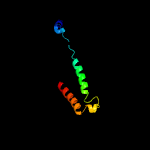
PDB 2i68 chain B
Region: 3 - 103
Aligned: 77
Modelled: 101
Confidence: 99.8%
Identity: 45%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 3 |
|
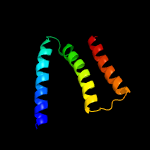
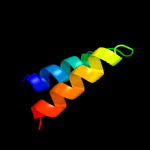
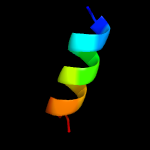
PDB 6oh2 chain A
Region: 4 - 103
Aligned: 98
Modelled: 100
Confidence: 98.1%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:cmp-sialic acid transporter;
PDBTitle: x-ray crystal structure of the mouse cmp-sialic acid transporter in2 complex with cmp, by lipidic cubic phase
Phyre2
| 4 |
|
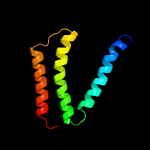
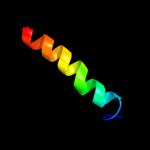
PDB 5i20 chain E
Region: 24 - 107
Aligned: 82
Modelled: 84
Confidence: 98.0%
Identity: 20%
PDB header:membrane protein
Chain: E: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein
Phyre2
| 5 |
|
PDB 6i1r chain A
Region: 21 - 103
Aligned: 81
Modelled: 83
Confidence: 97.9%
Identity: 5%
PDB header:membrane protein
Chain: A: PDB Molecule:cmp-sialic acid transporter 1;
PDBTitle: crystal structure of cmp bound cst in an outward facing conformation
Phyre2
| 6 |
|
PDB 5y79 chain A
Region: 29 - 104
Aligned: 74
Modelled: 76
Confidence: 97.8%
Identity: 16%
PDB header:transport protein
Chain: A: PDB Molecule:putative hexose phosphate translocator;
PDBTitle: crystal structure of the triose-phosphate/phosphate translocator in2 complex with 3-phosphoglycerate
Phyre2
| 7 |
|
PDB 5oge chain E
Region: 21 - 107
Aligned: 85
Modelled: 87
Confidence: 97.6%
Identity: 12%
PDB header:membrane protein
Chain: E: PDB Molecule:gdp-mannose transporter 1;
PDBTitle: crystal structure of a nucleotide sugar transporter
Phyre2
| 8 |
|
PDB 5i20 chain C
Region: 21 - 107
Aligned: 85
Modelled: 87
Confidence: 97.5%
Identity: 15%
PDB header:membrane protein
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein
Phyre2
| 9 |
|
PDB 4oo9 chain A
Region: 55 - 85
Aligned: 31
Modelled: 31
Confidence: 18.9%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:metabotropic glutamate receptor 5, lysozyme, metabotropic
PDBTitle: structure of the human class c gpcr metabotropic glutamate receptor 52 transmembrane domain in complex with the negative allosteric3 modulator mavoglurant
Phyre2
| 10 |
|
PDB 3mp7 chain B
Region: 85 - 101
Aligned: 17
Modelled: 17
Confidence: 16.2%
Identity: 29%
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
Phyre2
| 11 |
|
PDB 2lp1 chain A
Region: 77 - 100
Aligned: 24
Modelled: 24
Confidence: 9.0%
Identity: 33%
PDB header:membrane protein
Chain: A: PDB Molecule:c99;
PDBTitle: the solution nmr structure of the transmembrane c-terminal domain of2 the amyloid precursor protein (c99)
Phyre2
| 12 |
|
PDB 5xnm chain J
Region: 67 - 80
Aligned: 14
Modelled: 14
Confidence: 8.7%
Identity: 36%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
Phyre2
| 13 |
|
PDB 5azc chain A
Region: 27 - 101
Aligned: 75
Modelled: 75
Confidence: 7.9%
Identity: 19%
PDB header:transferase
Chain: A: PDB Molecule:prolipoprotein diacylglyceryl transferase;
PDBTitle: crystal structure of escherichia coli lgt in complex with2 phosphatidylglycerol
Phyre2
| 14 |
|
PDB 3jcu chain J
Region: 67 - 80
Aligned: 14
Modelled: 14
Confidence: 7.7%
Identity: 43%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
Phyre2
| 15 |
|
PDB 6e8w chain C
Region: 92 - 103
Aligned: 12
Modelled: 12
Confidence: 6.8%
Identity: 25%
PDB header:viral protein
Chain: C: PDB Molecule:envelope glycoprotein gp160;
PDBTitle: mper-tm domain of hiv-1 envelope glycoprotein (env)
Phyre2
| 16 |
|
PDB 5dir chain B
Region: 68 - 100
Aligned: 33
Modelled: 33
Confidence: 6.7%
Identity: 15%
PDB header:hydrolase
Chain: B: PDB Molecule:lipoprotein signal peptidase;
PDBTitle: membrane protein at 2.8 angstroms
Phyre2
| 17 |
|
PDB 5doq chain C
Region: 43 - 66
Aligned: 23
Modelled: 24
Confidence: 6.5%
Identity: 17%
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative membrane protein;
PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
Phyre2
| 18 |
|
PDB 5ir6 chain C
Region: 43 - 66
Aligned: 23
Modelled: 24
Confidence: 6.5%
Identity: 17%
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative membrane protein;
PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
Phyre2
| 19 |
|
PDB 6m97 chain A
Region: 13 - 80
Aligned: 60
Modelled: 68
Confidence: 6.3%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:chimera protein of high affinity copper uptake protein 1
PDBTitle: crystal structure of the high-affinity copper transporter ctr1
Phyre2
| 20 |
|
PDB 3a0h chain J
Region: 67 - 80
Aligned: 14
Modelled: 14
Confidence: 5.5%
Identity: 43%
PDB header:electron transport
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 21 |
|