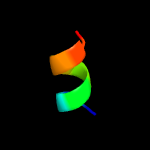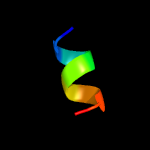| 1 |
|
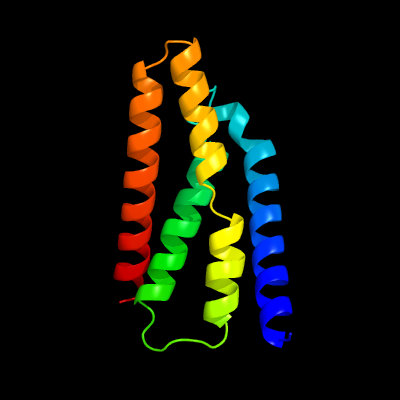
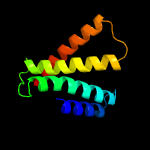
PDB 5a43 chain B
Region: 9 - 125
Aligned: 116
Modelled: 117
Confidence: 100.0%
Identity: 28%
PDB header:transport protein
Chain: B: PDB Molecule:putative fluoride ion transporter crcb;
PDBTitle: crystal structure of a dual topology fluoride ion channel.
Phyre2
| 2 |
|
PDB 5a40 chain C
Region: 10 - 125
Aligned: 113
Modelled: 113
Confidence: 100.0%
Identity: 31%
PDB header:transport protein
Chain: C: PDB Molecule:putative fluoride ion transporter crcb;
PDBTitle: crystal structure of a dual topology fluoride ion channel.
Phyre2
| 3 |
|
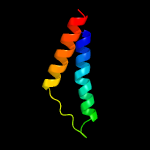
PDB 5edl chain A
Region: 23 - 123
Aligned: 100
Modelled: 101
Confidence: 25.1%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:putative hmp/thiamine permease protein ykoe;
PDBTitle: crystal structure of an s-component of ecf transporter
Phyre2
| 4 |
|
PDB 1m2z chain E
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 9.4%
Identity: 44%
PDB header:hormone/hormone activator
Chain: E: PDB Molecule:nuclear receptor coactivator 2;
PDBTitle: crystal structure of a dimer complex of the human glucocorticoid2 receptor ligand-binding domain bound to dexamethasone and a tif23 coactivator motif
Phyre2
| 5 |
|
PDB 2xok chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 8.9%
Identity: 25%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: refined structure of yeast f1c10 atpase complex to 3 a resolution
Phyre2
| 6 |
|
PDB 3gn8 chain C
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 8.0%
Identity: 44%
PDB header:hormone/hormone activator
Chain: C: PDB Molecule:nuclear receptor coactivator 2;
PDBTitle: x-ray crystal structure of ancgr2 in complex with2 dexamethasone
Phyre2
| 7 |
|
PDB 5lj7 chain B
Region: 12 - 62
Aligned: 51
Modelled: 51
Confidence: 7.5%
Identity: 18%
PDB header:transport protein
Chain: B: PDB Molecule:macrolide export atp-binding/permease protein macb;
PDBTitle: structure of aggregatibacter actinomycetemcomitans macb bound to atp2 (p21)
Phyre2
| 8 |
|
PDB 3gn8 chain E
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 7.5%
Identity: 44%
PDB header:hormone/hormone activator
Chain: E: PDB Molecule:nuclear receptor coactivator 2;
PDBTitle: x-ray crystal structure of ancgr2 in complex with2 dexamethasone
Phyre2
| 9 |
|
PDB 2w6h chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 7.2%
Identity: 38%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma, mitochondrial;
PDBTitle: low resolution structures of bovine mitochondrial f1-atpase during2 controlled dehydration: hydration state 4a.
Phyre2
| 10 |
|
PDB 2jdi chain G domain 1
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 38%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: ATP synthase (F1-ATPase), gamma subunit
Family: ATP synthase (F1-ATPase), gamma subunit
Phyre2
| 11 |
|
PDB 1m2z chain B
Region: 23 - 31
Aligned: 9
Modelled: 9
Confidence: 6.3%
Identity: 44%
PDB header:hormone/hormone activator
Chain: B: PDB Molecule:nuclear receptor coactivator 2;
PDBTitle: crystal structure of a dimer complex of the human glucocorticoid2 receptor ligand-binding domain bound to dexamethasone and a tif23 coactivator motif
Phyre2
| 12 |
|
PDB 1fs0 chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 25%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: ATP synthase (F1-ATPase), gamma subunit
Family: ATP synthase (F1-ATPase), gamma subunit
Phyre2
| 13 |
|
PDB 5lqx chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 25%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma subunit;
PDBTitle: structure of f-atpase from pichia angusta, state3
Phyre2
| 14 |
|
PDB 6foc chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 6.0%
Identity: 50%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: f1-atpase from mycobacterium smegmatis
Phyre2
| 15 |
|
PDB 5zwl chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 50%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: crystal structure of the gamma - epsilon complex of photosynthetic2 cyanobacterial f1-atpase
Phyre2
| 16 |
|
PDB 2qe7 chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 5.8%
Identity: 38%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase subunit gamma;
PDBTitle: crystal structure of the f1-atpase from the thermoalkaliphilic2 bacterium bacillus sp. ta2.a1
Phyre2
| 17 |
|
PDB 5dn6 chain G
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 5.7%
Identity: 25%
PDB header:hydrolase
Chain: G: PDB Molecule:atp synthase gamma chain;
PDBTitle: atp synthase from paracoccus denitrificans
Phyre2
| 18 |
|
PDB 3oaa chain O
Region: 75 - 82
Aligned: 8
Modelled: 8
Confidence: 5.7%
Identity: 25%
PDB header:hydrolase/transport protein
Chain: O: PDB Molecule:atp synthase gamma chain;
PDBTitle: structure of the e.coli f1-atp synthase inhibited by subunit epsilon
Phyre2