| 1 | c3sdoB_
|
|
 |
99.9 |
18 |
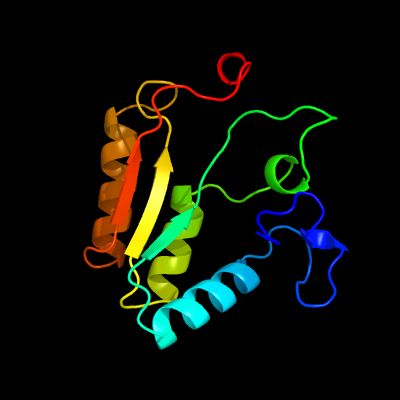
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
| 2 | c1tvlA_
|
|
 |
99.9 |
15 |
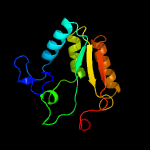
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 3 | d1tvla_
|
|
 |
99.9 |
15 |
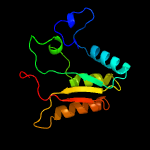
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 4 | c5tlcA_
|
|
 |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
| 5 | c2b81D_
|
|
 |
99.9 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
|
| 6 | c3raoB_
|
|
 |
99.9 |
29 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
| 7 | c5w4zA_
|
|
 |
99.9 |
19 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
| 8 | c3b9nB_
|
|
 |
99.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 9 | d1lucb_
|
|
 |
99.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 10 | c2i7gA_
|
|
 |
99.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
| 11 | c5dqpA_
|
|
 |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 12 | d1luca_
|
|
 |
99.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 13 | d1rhca_
|
|
 |
99.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 14 | c2wgkA_
|
|
 |
99.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
| 15 | c6friD_
|
|
 |
99.9 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
| 16 | d1ezwa_
|
|
 |
99.9 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 17 | c5wanA_
|
|
 |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
|
| 18 | c6ak1B_
|
|
 |
99.9 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
|
| 19 | c3c8nB_
|
|
 |
99.9 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 20 | c1z69D_
|
|
 |
99.9 |
25 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
| 21 | d1f07a_ |
|
not modelled |
99.9 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
| 22 | d1nqka_ |
|
not modelled |
99.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
| 23 | c3qy6A_ |
|
not modelled |
67.1 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 24 | d1oy0a_ |
|
not modelled |
53.1 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 25 | d1oyaa_ |
|
not modelled |
44.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 26 | c2ze3A_ |
|
not modelled |
44.8 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 27 | c6ei9A_ |
|
not modelled |
43.9 |
23 |
PDB header:flavoprotein
Chain: A: PDB Molecule:trna-dihydrouridine synthase b;
PDBTitle: crystal structure of e. coli trna-dihydrouridine synthase b (dusb)
|
|
|
| 28 | c3bwwA_ |
|
not modelled |
43.6 |
13 |
PDB header:metal binding protein
Chain: A: PDB Molecule:protein of unknown function duf692/cog3220;
PDBTitle: crystal structure of a duf692 family protein (hs_1138) from2 haemophilus somnus 129pt at 2.20 a resolution
|
|
|
| 29 | c1zlpA_ |
|
not modelled |
43.4 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 30 | c5dxxA_ |
|
not modelled |
43.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:artemisinic aldehyde delta(11(13)) reductase;
PDBTitle: crystal structure of dbr2
|
|
|
| 31 | d1e8ca2 |
|
not modelled |
41.9 |
13 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
| 32 | c3e96B_ |
|
not modelled |
41.8 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 33 | d1z41a1 |
|
not modelled |
41.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 34 | c4b5nA_ |
|
not modelled |
40.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: crystal structure of oxidized shewanella yellow enzyme 4 (sye4)
|
|
|
| 35 | c3b0vD_ |
|
not modelled |
40.6 |
23 |
PDB header:oxidoreductase/rna
Chain: D: PDB Molecule:trna-dihydrouridine synthase;
PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
|
|
|
| 36 | c3dcpB_ |
|
not modelled |
40.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of the putative histidinol phosphatase hisk from2 listeria monocytogenes. northeast structural genomics consortium3 target lmr141.
|
|
|
| 37 | c4lsbA_ |
|
not modelled |
40.1 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:lyase/mutase;
PDBTitle: crystal structure of a putative lyase/mutase from burkholderia2 cenocepacia j2315
|
|
|
| 38 | c2h90A_ |
|
not modelled |
39.2 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
|
|
| 39 | c3lyeA_ |
|
not modelled |
38.7 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 40 | d1s2wa_ |
|
not modelled |
38.5 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 41 | c2wjeA_ |
|
not modelled |
38.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
|
|
| 42 | c4a3uB_ |
|
not modelled |
37.8 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 43 | c4tmcB_ |
|
not modelled |
37.3 |
20 |
PDB header:flavoprotein
Chain: B: PDB Molecule:old yellow enzyme;
PDBTitle: crystal structure of old yellow enzyme from candida macedoniensis2 aku4588 complexed with p-hydroxybenzaldehyde
|
|
|
| 44 | d1vjia_ |
|
not modelled |
36.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 45 | d1gwja_ |
|
not modelled |
36.6 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 46 | c2qiwA_ |
|
not modelled |
35.8 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 47 | c3l5aA_ |
|
not modelled |
35.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
|
|
| 48 | c3fa4D_ |
|
not modelled |
35.0 |
33 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 49 | c2gq8A_ |
|
not modelled |
34.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
|
|
| 50 | d1muma_ |
|
not modelled |
34.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 51 | c2yb1A_ |
|
not modelled |
34.7 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
|
|
|
| 52 | d1q45a_ |
|
not modelled |
34.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 53 | d1vyra_ |
|
not modelled |
34.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 54 | c5n2pA_ |
|
not modelled |
33.2 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 55 | c4jicB_ |
|
not modelled |
32.7 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gtn reductase;
PDBTitle: glycerol trinitrate reductase nera from agrobacterium radiobacter
|
|
|
| 56 | c2fptA_ |
|
not modelled |
32.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase, mitochondrial;
PDBTitle: dual binding mode of a novel series of dhodh inhibitors
|
|
|
| 57 | c4rnxA_ |
|
not modelled |
30.6 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase 1;
PDBTitle: k154 circular permutation of old yellow enzyme
|
|
|
| 58 | d1icpa_ |
|
not modelled |
30.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 59 | c3d0cB_ |
|
not modelled |
30.2 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 60 | c3gr7A_ |
|
not modelled |
29.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 61 | c3eooL_ |
|
not modelled |
28.9 |
27 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 62 | c3b8iF_ |
|
not modelled |
28.8 |
40 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 63 | c5uncB_ |
|
not modelled |
28.8 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoenolpyruvate phosphomutase;
PDBTitle: the crystal structure of phosphoenolpyruvate phosphomutase from2 streptomyces platensis subsp. rosaceus
|
|
|
| 64 | d1m3ua_ |
|
not modelled |
28.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 65 | c4rnvD_ |
|
not modelled |
28.3 |
10 |
PDB header:oxidoreductase/inhibitor
Chain: D: PDB Molecule:nadph dehydrogenase 1;
PDBTitle: g303 circular permutation of old yellow enzyme with the inhibitor p-2 hydroxybenzaldehyde
|
|
|
| 66 | c3kruC_ |
|
not modelled |
28.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
|
|
|
| 67 | c3gkaB_ |
|
not modelled |
28.0 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
|
|
| 68 | d1o5ka_ |
|
not modelled |
27.7 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 69 | d1djqa1 |
|
not modelled |
27.6 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 70 | c6ahuJ_ |
|
not modelled |
27.6 |
19 |
PDB header:hydrolase/rna
Chain: J: PDB Molecule:ribonuclease p protein subunit p30;
PDBTitle: cryo-em structure of human ribonuclease p with mature trna
|
|
|
| 71 | c3na8A_ |
|
not modelled |
27.6 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
|
|
|
| 72 | c4qnwA_ |
|
not modelled |
26.6 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chanoclavine-i aldehyde reductase;
PDBTitle: crystal structure of easa, an old yellow enzyme from aspergillus2 fumigatus
|
|
|
| 73 | c6daqA_ |
|
not modelled |
26.2 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:phdj;
PDBTitle: phdj bound to substrate intermediate
|
|
|
| 74 | c4df2A_ |
|
not modelled |
26.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: p. stipitis oye2.6 complexed with p-chlorophenol
|
|
|
| 75 | c3w9zA_ |
|
not modelled |
25.4 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna-dihydrouridine synthase c;
PDBTitle: crystal structure of dusc
|
|
|
| 76 | c4ot7A_ |
|
not modelled |
24.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of a variant of ncr from zymomonas mobilis
|
|
|
| 77 | d3d1ma1 |
|
not modelled |
24.6 |
25 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
|
|
| 78 | d1m65a_ |
|
not modelled |
24.5 |
14 |
Fold:7-stranded beta/alpha barrel
Superfamily:PHP domain-like
Family:PHP domain |
|
|
| 79 | c6k0aC_ |
|
not modelled |
22.9 |
0 |
PDB header:rna binding protein/rna
Chain: C: PDB Molecule:ribonuclease p protein component 3;
PDBTitle: cryo-em structure of an archaeal ribonuclease p
|
|
|
| 80 | d1ps9a1 |
|
not modelled |
22.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 81 | c3ih1A_ |
|
not modelled |
22.8 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 82 | c3m1nB_ |
|
not modelled |
22.5 |
25 |
PDB header:signaling protein
Chain: B: PDB Molecule:sonic hedgehog protein;
PDBTitle: crystal structure of human sonic hedgehog n-terminal domain
|
|
|
| 83 | c3bh1A_ |
|
not modelled |
22.5 |
45 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
|
|
| 84 | c3i4eA_ |
|
not modelled |
21.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:isocitrate lyase;
PDBTitle: crystal structure of isocitrate lyase from burkholderia2 pseudomallei
|
|
|
| 85 | c6agzA_ |
|
not modelled |
20.8 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:old yellow enzyme;
PDBTitle: crystal structure of old yellow enzyme from pichia sp. aku4542
|
|
|
| 86 | d1o66a_ |
|
not modelled |
20.7 |
37 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 87 | c6mywA_ |
|
not modelled |
20.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: gluconobacter ene-reductase (gluer) mutant - t36a
|
|
|
| 88 | d2ibge1 |
|
not modelled |
20.4 |
25 |
Fold:Hedgehog/DD-peptidase
Superfamily:Hedgehog/DD-peptidase
Family:Hedgehog (development protein), N-terminal signaling domain |
|
|
| 89 | c6de6B_ |
|
not modelled |
20.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: 2.1 a resolution structure of histamine dehydrogenase from rhizobium2 sp. 4-9
|
|
|
| 90 | c1ydnA_ |
|
not modelled |
20.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
|
|
| 91 | c5epdA_ |
|
not modelled |
19.8 |
20 |
PDB header:oxidorecctase
Chain: A: PDB Molecule:glycerol trinitrate reductase;
PDBTitle: crystal structure of glycerol trinitrate reductase xdpb from2 agrobacterium sp. r89-1 (apo form)
|
|
|
| 92 | d1yxya1 |
|
not modelled |
19.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 93 | c2w9mB_ |
|
not modelled |
19.3 |
24 |
PDB header:dna replication
Chain: B: PDB Molecule:polymerase x;
PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
|
|
|
| 94 | c3eb2A_ |
|
not modelled |
18.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
|
|
| 95 | c3e0fA_ |
|
not modelled |
18.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent phosphoesterase;
PDBTitle: crystal structure of a putative metal-dependent phosphoesterase2 (bad_1165) from bifidobacterium adolescentis atcc 15703 at 2.40 a3 resolution
|
|
|
| 96 | c3vsjA_ |
|
not modelled |
18.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit;
PDBTitle: crystal structure of 1,6-apd (2-animophenol-1,6-dioxygenase) complexed2 with intermediate products
|
|
|
| 97 | c3hf3A_ |
|
not modelled |
18.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
|
|
| 98 | c5ocsB_ |
|
not modelled |
17.8 |
22 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 99 | c4gx9A_ |
|
not modelled |
17.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii subunit epsilon,dna polymerase iii
PDBTitle: crystal structure of a dna polymerase iii alpha-epsilon chimera
|
|
|