1 c3bk6C_
99.9
8
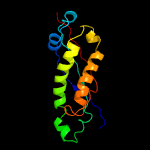
PDB header: membrane proteinChain: C: PDB Molecule: ph stomatin;PDBTitle: crystal structure of a core domain of stomatin from2 pyrococcus horikoshii
2 d1wina_
99.7
8
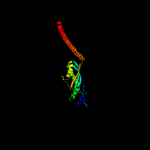
Fold: EF-Ts domain-likeSuperfamily: Band 7/SPFH domainFamily: Band 7/SPFH domain
3 c4fvjB_
99.7
9
PDB header: membrane proteinChain: B: PDB Molecule: stomatin;PDBTitle: spfh domain of the mouse stomatin (crystal form 2)
4 c2rpbA_
99.6
13
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical membrane protein;PDBTitle: the solution structure of membrane protein
5 c2zv4O_
97.7
12
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
6 c2qzvB_
95.5
13
PDB header: structural proteinChain: B: PDB Molecule: major vault protein;PDBTitle: draft crystal structure of the vault shell at 9 angstroms2 resolution
7 c2kk7A_
79.0
21
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit e;PDBTitle: nmr solution structure of the n terminal domain of subunit e2 (e1-52) of a1ao atp synthase from methanocaldococcus3 jannaschii
8 c3k5bB_
37.8
12
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase, subunit (vapc-therm);PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
9 c6ahqM_
34.7
13
PDB header: motor proteinChain: M: PDB Molecule: flagellar protein flil;PDBTitle: structure of the 40-167 fragment of flil
10 c2k6iA_
25.3
21
PDB header: structural proteinChain: A: PDB Molecule: uncharacterized protein mj0223;PDBTitle: the domain features of the peripheral stalk subunit h of the2 methanogenic a1ao atp synthase and the nmr solution3 structure of h1-47
11 d1nkta1
24.0
37
Fold: Pre-protein crosslinking domain of SecASuperfamily: Pre-protein crosslinking domain of SecAFamily: Pre-protein crosslinking domain of SecA
12 d1tf5a1
21.8
33
Fold: Pre-protein crosslinking domain of SecASuperfamily: Pre-protein crosslinking domain of SecAFamily: Pre-protein crosslinking domain of SecA
13 c3wvfA_
19.8
19
PDB header: chaperoneChain: A: PDB Molecule: membrane protein insertase yidc;PDBTitle: crystal structure of yidc from escherichia coli
14 d2diga1
19.5
54
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain
15 c3k5bE_
18.7
16
PDB header: hydrolaseChain: E: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
16 c6mctC_
17.9
26
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
17 c6mctL_
17.9
26
PDB header: de novo proteinChain: L: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
18 c6mctJ_
17.9
26
PDB header: de novo proteinChain: J: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
19 c6mctF_
17.9
26
PDB header: de novo proteinChain: F: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
20 c6mq2D_
17.9
26
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
21 c6mctO_
not modelled
17.9
26
PDB header: de novo proteinChain: O: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
22 c6mctE_
not modelled
17.9
26
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
23 c6mpwA_
not modelled
17.9
26
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
24 c6mctG_
not modelled
17.9
26
PDB header: de novo proteinChain: G: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
25 c6mctA_
not modelled
17.9
26
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
26 c6mctI_
not modelled
17.9
26
PDB header: de novo proteinChain: I: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
27 c6mctH_
not modelled
17.9
26
PDB header: de novo proteinChain: H: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
28 c6mctK_
not modelled
17.9
26
PDB header: de novo proteinChain: K: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
29 c6mctB_
not modelled
17.9
26
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
30 c6mctM_
not modelled
17.9
26
PDB header: de novo proteinChain: M: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
31 c6mctN_
not modelled
17.9
26
PDB header: de novo proteinChain: N: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
32 c6mctD_
not modelled
17.9
26
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: a designed pentameric membrane protein stabilized by van der waals2 interaction
33 c6mpwD_
not modelled
17.3
26
PDB header: de novo proteinChain: D: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
34 c6mq2B_
not modelled
17.3
26
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
35 c6mpwB_
not modelled
17.3
26
PDB header: de novo proteinChain: B: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
36 c6mpwC_
not modelled
17.3
26
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
37 c6mq2E_
not modelled
17.3
26
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
38 c6mq2A_
not modelled
17.3
26
PDB header: de novo proteinChain: A: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
39 c6mq2C_
not modelled
17.3
26
PDB header: de novo proteinChain: C: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-2
40 c6mpwE_
not modelled
17.3
26
PDB header: de novo proteinChain: E: PDB Molecule: mini-evgl membrane protein;PDBTitle: de novo design of membrane protein--mini-evgl membrane protein, c22212 form-1
41 c4tt1A_
not modelled
14.9
9
PDB header: hydrolaseChain: A: PDB Molecule: deneddylase;PDBTitle: crystal structure of fragment 1600-1733 of hsv1 ul36, native
42 c4pj0l_
not modelled
14.6
3
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
43 c4pj0L_
not modelled
14.6
3
PDB header: oxidoreductase, electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of t.elongatus photosystem ii, rows of dimers crystal2 packing
44 c4tnhL_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
45 c4tnjl_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
46 c3wu2l_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
47 c4fbyd_
not modelled
14.5
3
PDB header: photosynthesisChain: D: PDB Molecule: photosystem ii d2 protein;PDBTitle: fs x-ray diffraction of photosystem ii
48 c4tniL_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
49 c4fbyL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: fs x-ray diffraction of photosystem ii
50 c3bz2L_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 2 of 2). this2 file contains second monomer of psii dimer
51 c1s5ll_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
52 c4tnjL_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the 2nd illumination2 (2f) at 4.5 a resolution
53 c4tnhl_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii in the dark state at 4.9 a2 resolution
54 c4tnil_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 500 ms after the third2 illumination at 4.6 a resolution
55 c4ub8L_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
56 c4ub6l_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
57 c4tnkl_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
58 c2axtl_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
59 c4tnkL_
not modelled
14.5
3
PDB header: electron transport,photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt xfel structure of photosystem ii 250 microsec after the third2 illumination at 5.2 a resolution
60 c3wu2L_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure analysis of photosystem ii complex
61 c4ub6L_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
62 c4ub8l_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
63 c1s5lL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
64 c3prqL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
65 c3bz1L_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 1 of 2). this2 file contains first monomer of psii dimer
66 c3kziL_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
67 c2axtL_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center l protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
68 d2axtl1
not modelled
14.5
3
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein L, PsbLFamily: PsbL-like
69 c3arcL_
not modelled
14.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
70 c3a0bl_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
71 c4rvyL_
not modelled
14.5
3
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
72 c3prrL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
73 c5e7cl_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
74 c4ixqL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
75 c4il6L_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
76 c4ixrl_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
77 c4ixrL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, first illuminated state
78 c5e7cL_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: macromolecular diffractive imaging using imperfect crystals - bragg2 data
79 c4il6l_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: structure of sr-substituted photosystem ii
80 c4ixql_
not modelled
14.5
3
PDB header: photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: rt fs x-ray diffraction of photosystem ii, dark state
81 c4rvyl_
not modelled
14.5
3
PDB header: oxidoreductaseChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: serial time resolved crystallography of photosystem ii using a2 femtosecond x-ray laser. the s state after two flashes (s3)
82 c3a0hl_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
83 c3a0bL_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of br-substituted photosystem ii complex
84 c3a0hL_
not modelled
14.5
3
PDB header: electron transportChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of i-substituted photosystem ii complex
85 c4dl0J_
not modelled
11.2
8
PDB header: hydrolaseChain: J: PDB Molecule: v-type proton atpase subunit e;PDBTitle: crystal structure of the heterotrimeric egchead peripheral stalk2 complex of the yeast vacuolar atpase
86 c5a8fA_
not modelled
10.9
14
PDB header: viral proteinChain: A: PDB Molecule: human saffold virus-3 vp1;PDBTitle: structure and genome release mechanism of human cardiovirus saffold2 virus-3
87 c4or2A_
not modelled
10.7
8
PDB header: signaling proteinChain: A: PDB Molecule: soluble cytochrome b562, metabotropic glutamate receptor 1;PDBTitle: human class c g protein-coupled metabotropic glutamate receptor 1 in2 complex with a negative allosteric modulator
88 c3arcl_
not modelled
10.5
3
PDB header: electron transport, photosynthesisChain: L: PDB Molecule: photosystem ii reaction center protein l;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
89 c2jp3A_
not modelled
10.1
8
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
90 c2jo1A_
not modelled
9.6
12
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
91 c2digA_
not modelled
9.3
54
PDB header: dna binding proteinChain: A: PDB Molecule: lamin-b receptor;PDBTitle: solusion structure of the todor domain of human lamin-b2 receptor
92 c2zxeG_
not modelled
8.9
27
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
93 c2mkvA_
not modelled
8.6
19
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
94 c4dl0G_
not modelled
8.5
26
PDB header: hydrolaseChain: G: PDB Molecule: v-type proton atpase subunit g;PDBTitle: crystal structure of the heterotrimeric egchead peripheral stalk2 complex of the yeast vacuolar atpase
95 c1wd6B_
not modelled
8.4
38
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein ydhr;PDBTitle: crystal structure of jw1657 from escherichia coli
96 d1eg2a_
not modelled
8.2
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Type II DNA methylase
97 c6nbxG_
not modelled
7.4
18
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: t.elongatus ndh (data-set 2)
98 c6c6lO_
not modelled
7.3
15
PDB header: membrane proteinChain: O: PDB Molecule: v-type proton atpase subunit f;PDBTitle: yeast vacuolar atpase vo in lipid nanodisc
99 c6n52B_
not modelled
7.0
12
PDB header: membrane proteinChain: B: PDB Molecule: metabotropic glutamate receptor 5;PDBTitle: metabotropic glutamate receptor 5 apo form