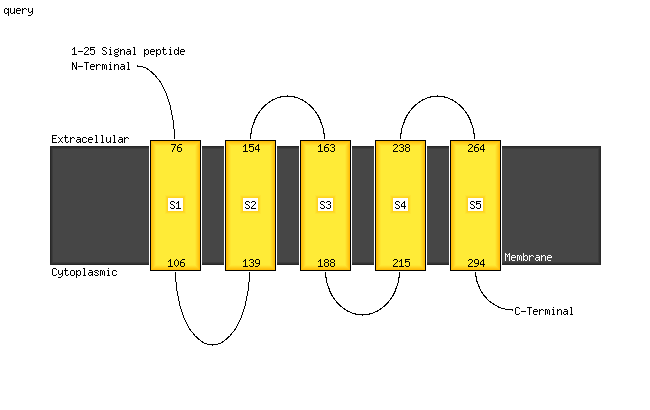
1 c5wwoB_
40.8
27
PDB header: rna binding proteinChain: B: PDB Molecule: essential nuclear protein 1;PDBTitle: crystal structure of enp1
2 d2enda_
37.0
35
Fold: T4 endonuclease VSuperfamily: T4 endonuclease VFamily: T4 endonuclease V
3 c2l9uB_
36.5
41
PDB header: membrane proteinChain: B: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
4 c2l9uA_
36.5
41
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
5 c4cv5D_
27.3
21
PDB header: cell cycleChain: D: PDB Molecule: protein caf40;PDBTitle: yeast not1 cn9bd-caf40 complex
6 c5doqC_
21.9
48
PDB header: oxidoreductaseChain: C: PDB Molecule: putative membrane protein;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
7 c5ir6C_
21.9
48
PDB header: oxidoreductaseChain: C: PDB Molecule: putative membrane protein;PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
8 c5tvoB_
20.2
25
PDB header: lyaseChain: B: PDB Molecule: s-adenosylmethionine decarboxylase proenzyme;PDBTitle: crystal structure of trypanosoma brucei adometdc-delta26 monomer
9 c5tvfC_
18.5
30
PDB header: lyaseChain: C: PDB Molecule: s-adenosylmethionine decarboxylase beta chain;PDBTitle: crystal structure of trypanosoma brucei adometdc/prozyme heterodimer2 in complex with inhibitor cgp 40215
10 c2voyB_
16.5
22
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calcium atpase 1;PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
11 c4r0cB_
16.4
17
PDB header: membrane proteinChain: B: PDB Molecule: abgt putative transporter family;PDBTitle: crystal structure of the alcanivorax borkumensis ydah transporter2 reveals an unusual topology
12 d1nexa2
16.2
19
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain
13 c4gl6B_
15.6
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a duf5037 family protein (rumgna_01148) from2 ruminococcus gnavus atcc 29149 at 2.55 a resolution
14 c1i7mD_
15.0
44
PDB header: lyaseChain: D: PDB Molecule: s-adenosylmethionine decarboxylase beta chain;PDBTitle: human s-adenosylmethionine decarboxylase with covalently bound2 pyruvoyl group and complexed with 4-amidinoindan-1-one-2'-3 amidinohydrazone
15 c6g4ww_
14.0
24
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s15a;PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state a
16 c4zr0A_
13.5
27
PDB header: oxidoreductaseChain: A: PDB Molecule: ceramide very long chain fatty acid hydroxylase scs7;PDBTitle: full length scs7p (only hydroxylase domain visible)
17 c1w8xP_
12.5
60
PDB header: virusChain: P: PDB Molecule: protein p16;PDBTitle: structural analysis of prd1
18 d2c9wc1
10.4
18
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain
19 c5j6vA_
9.9
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: hylin-d;PDBTitle: nmr structures of hylin-a1 analogs: hylin-d
20 c5j6wA_
9.5
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: hylin-k;PDBTitle: nmr structures of hylin-a1 analogs: hylin-k
21 d1kf6d_
not modelled
8.6
20
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
22 c6ig0H_
not modelled
8.6
50
PDB header: rna binding proteinChain: H: PDB Molecule: type iii-a crispr-associated ramp protein csm5;PDBTitle: type iii-a csm complex, cryo-em structure of csm-ctr1, atp bound
23 d1urfa_
not modelled
7.3
19
Fold: Long alpha-hairpinSuperfamily: HR1 repeatFamily: HR1 repeat
24 c6musF_
not modelled
7.3
50
PDB header: rna binding protein/rnaChain: F: PDB Molecule: uncharacterized protein csm5;PDBTitle: cryo-em structure of larger csm-crrna-target rna ternary complex in2 type iii-a crispr-cas system
25 c5lqzW_
not modelled
6.6
12
PDB header: hydrolaseChain: W: PDB Molecule: atp synthase subunit d;PDBTitle: structure of f-atpase from pichia angusta, state1
26 c5lqxW_
not modelled
6.6
12
PDB header: hydrolaseChain: W: PDB Molecule: atp synthase subunit d;PDBTitle: structure of f-atpase from pichia angusta, state3
27 c5lqyW_
not modelled
6.6
12
PDB header: hydrolaseChain: W: PDB Molecule: atp synthase subunit d;PDBTitle: structure of f-atpase from pichia angusta, in state2
28 c2zjsE_
not modelled
6.5
23
PDB header: protein transport/immune systemChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: crystal structure of secye translocon from thermus thermophilus with a2 fab fragment
29 c2zqpE_
not modelled
6.5
23
PDB header: protein transportChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: crystal structure of secye translocon from thermus2 thermophilus
30 c1msvB_
not modelled
6.4
44
PDB header: lyaseChain: B: PDB Molecule: s-adenosylmethionine decarboxylase proenzyme;PDBTitle: the s68a s-adenosylmethionine decarboxylase proenzyme2 processing mutant.
31 c5gtuB_
not modelled
6.4
57
PDB header: hydrolaseChain: B: PDB Molecule: tbc1 domain family member 5;PDBTitle: structural and mechanistic insights into regulation of the retromer2 coat by tbc1d5
32 d1fs1b2
not modelled
6.3
23
Fold: POZ domainSuperfamily: POZ domainFamily: BTB/POZ domain
33 d1jl0a_
not modelled
6.1
44
Fold: S-adenosylmethionine decarboxylaseSuperfamily: S-adenosylmethionine decarboxylaseFamily: S-adenosylmethionine decarboxylase
34 c4px7A_
not modelled
6.0
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylglycerophosphatase;PDBTitle: crystal structure of lipid phosphatase e. coli pgpb
35 d1lgha_
not modelled
5.8
36
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits
36 c4fe1I_
not modelled
5.8
33
PDB header: photosynthesisChain: I: PDB Molecule: photosystem i reaction center subunit viii;PDBTitle: improving the accuracy of macromolecular structure refinement at 7 a2 resolution
37 d1jb0i_
not modelled
5.8
33
Fold: Single transmembrane helixSuperfamily: Subunit VIII of photosystem I reaction centre, PsaIFamily: Subunit VIII of photosystem I reaction centre, PsaI
38 c5wujA_
not modelled
5.6
22
PDB header: motor proteinChain: A: PDB Molecule: flagellar m-ring protein;PDBTitle: crystal structure of flif-flig complex from h. pylori
39 c2m55B_
not modelled
5.6
33
PDB header: calcium binding protein/protein fibrilChain: B: PDB Molecule: alpha-synuclein;PDBTitle: nmr structure of the complex of an n-terminally acetylated alpha-2 synuclein peptide with calmodulin
40 c6bcdB_
not modelled
5.5
23
PDB header: replicationChain: B: PDB Molecule: dna polymerase zeta catalytic subunit;PDBTitle: crystal structure of rev7-k44a/r124a/a135d in complex with rev3-rbm22 (residues 1988-2014)
41 d2bapa1
not modelled
5.4
18
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Diap1 N-terninal region-like
42 c5w7lA_
not modelled
5.3
15
PDB header: membrane proteinChain: A: PDB Molecule: n,n'-diacetylbacilliosaminyl-1-phosphate transferase;PDBTitle: structure of campylobacter concisus pglc i57m/q175m variant
43 c6bc8B_
not modelled
5.2
23
PDB header: replicationChain: B: PDB Molecule: dna polymerase zeta catalytic subunit;PDBTitle: crystal structure of rev7-r124a/rev3-rbm2 (residues 1988-2014) complex