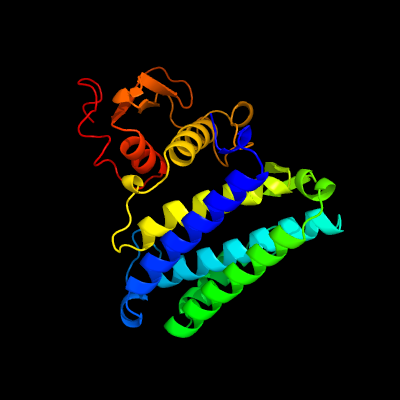
| 1 | c2nsfA_
|
|
 |
100.0 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:hypothetical protein cgl3021;
PDBTitle: crystal structure of the mycothiol-dependent maleylpyruvate isomerase
|
|
|
|
| 2 | d2nsfa1
|
|
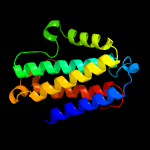 |
100.0 |
14 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:Maleylpyruvate isomerase-like |
|
|
|
| 3 | c5civA_
|
|
 |
99.6 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:sibling bacteriocin;
PDBTitle: sibling lethal factor precursor - dfsb
|
|
|
|
| 4 | c5cogB_
|
|
 |
99.5 |
15 |
PDB header:unknown function
Chain: B: PDB Molecule:irc4;
PDBTitle: crystal structure of yeast irc4
|
|
|
|
| 5 | d1rxqa_
|
|
 |
99.4 |
15 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:YfiT-like putative metal-dependent hydrolases |
|
|
|
| 6 | c4n6cB_
|
|
 |
99.4 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the b1rzq2 protein from streptococcus pneumoniae.2 northeast structural genomics consortium (nesg) target spr36.
|
|
|
|
| 7 | c2rd9C_
|
|
 |
99.2 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:bh0186 protein;
PDBTitle: crystal structure of a putative yfit-like metal-dependent hydrolase2 (bh0186) from bacillus halodurans c-125 at 2.30 a resolution
|
|
|
|
| 8 | c6iz2A_
|
|
 |
99.2 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:dinb/yfit family protein;
PDBTitle: crystal structure of dinb/yfit protein dr0053 from d. radiodurans r1
|
|
|
|
| 9 | c5cofA_
|
|
 |
99.2 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterised protein q1r1x2 from escherichia2 coli uti89
|
|
|
|
| 10 | c3cexB_
|
|
 |
99.1 |
8 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the conserved protein of locus ef_3021 from2 enterococcus faecalis
|
|
|
|
| 11 | c6anrA_
|
|
 |
99.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:colibactin self-protection protein clbs;
PDBTitle: crystal structure of a self resistance protein clbs from colibactin2 biosynthetic gene cluster
|
|
|
|
| 12 | d2ou6a1
|
|
 |
99.0 |
15 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:DinB-like |
|
|
|
| 13 | c5cqvB_
|
|
 |
98.9 |
9 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein q8dwv2 from streptococcus2 agalactiae
|
|
|
|
| 14 | c3dkaA_
|
|
 |
98.8 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:dinb-like protein;
PDBTitle: crystal structure of a dinb-like protein (yjoa, bsu12410) from2 bacillus subtilis at 2.30 a resolution
|
|
|
|
| 15 | c2yqyB_
|
|
 |
98.8 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein ttha0303;
PDBTitle: crystal structure of tt2238, a four-helix bundle protein
|
|
|
|
| 16 | d2p1aa1
|
|
 |
98.8 |
13 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:DinB-like |
|
|
|
| 17 | c3di5A_
|
|
 |
98.7 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:dinb-like protein;
PDBTitle: crystal structure of a dinb-like protein (bce_4655) from bacillus2 cereus atcc 10987 at 2.01 a resolution
|
|
|
|
| 18 | d2hkva1
|
|
 |
98.6 |
13 |
Fold:DinB/YfiT-like putative metalloenzymes
Superfamily:DinB/YfiT-like putative metalloenzymes
Family:DinB-like |
|
|
|
| 19 | c3e4xB_
|
|
 |
98.5 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:apc36150;
PDBTitle: crystal structure of putative metal-dependent hydrolases2 apc36150
|
|
|
|
| 20 | c2qe9B_
|
|
 |
98.4 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized protein yiza;
PDBTitle: crystal structure of a putative metal-dependent hydrolase (yiza,2 bsu10800) from bacillus subtilis at 1.90 a resolution
|
|
|
|
| 21 | c6h6pA_ |
|
not modelled |
97.7 |
15 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:ubiquinone biosynthesis protein ubij;
PDBTitle: ubij-scp2 ubiquinone synthesis protein
|
|
|
| 22 | c3bdqB_ |
|
not modelled |
97.7 |
19 |
PDB header:lipid transport
Chain: B: PDB Molecule:sterol carrier protein 2-like 2;
PDBTitle: room tempreture crystal structure of sterol carrier protein-2 2 like-2
|
|
|
| 23 | d2cfua1 |
|
not modelled |
97.5 |
12 |
Fold:SCP-like
Superfamily:SCP-like
Family:Alkylsulfatase C-terminal domain-like |
|
|
| 24 | d1ikta_ |
|
not modelled |
97.2 |
15 |
Fold:SCP-like
Superfamily:SCP-like
Family:Sterol carrier protein, SCP |
|
|
| 25 | d1pz4a_ |
|
not modelled |
97.1 |
15 |
Fold:SCP-like
Superfamily:SCP-like
Family:Sterol carrier protein, SCP |
|
|
| 26 | d1c44a_ |
|
not modelled |
97.1 |
14 |
Fold:SCP-like
Superfamily:SCP-like
Family:Sterol carrier protein, SCP |
|
|
| 27 | c4jgxB_ |
|
not modelled |
96.8 |
13 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:fatty acid-binding protein;
PDBTitle: the structure of sterol carrier protein 2 from the yeast yarrowia2 lipolytica
|
|
|
| 28 | c5wk0A_ |
|
not modelled |
96.8 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:damage-inducible protein dinb;
PDBTitle: crystal structure of the bacillithiol transferase bsta from2 staphylococcus aureus.
|
|
|
| 29 | c4ueiA_ |
|
not modelled |
96.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:sterol carrier protein 2/3-oxoacyl-coa thiolase;
PDBTitle: solution structure of the sterol carrier protein domain 22 of helicoverpa armigera
|
|
|
| 30 | c3bkrA_ |
|
not modelled |
96.5 |
18 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:sterol carrier protein-2 like-3;
PDBTitle: crystal structure of sterol carrier protein-2 like-3 (scp2-2 l3) from aedes aegypti
|
|
|
| 31 | c3bn8A_ |
|
not modelled |
95.3 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:putative sterol carrier protein 2;
PDBTitle: crystal structure of a putative sterol carrier protein type 2 (af1534)2 from archaeoglobus fulgidus dsm 4304 at 2.11 a resolution
|
|
|
| 32 | c4pdxB_ |
|
not modelled |
95.0 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alkyl/aryl-sulfatase yjcs;
PDBTitle: crystal structure of escherchia coli uncharacterized protein yjcs
|
|
|
| 33 | c4nurB_ |
|
not modelled |
87.4 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:psdsa;
PDBTitle: crystal structure of thermostable alkylsulfatase sdsap from2 pseudomonas sp. s9
|
|
|
| 34 | c2cfuA_ |
|
not modelled |
87.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:sdsa1;
PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
|
|
|
| 35 | c2yheD_ |
|
not modelled |
87.0 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:sec-alkyl sulfatase;
PDBTitle: structure determination of the stereoselective inverting sec-2 alkylsulfatase pisa1 from pseudomonas sp.
|
|
|
| 36 | c2qnlA_ |
|
not modelled |
85.4 |
9 |
PDB header:signaling protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative dna damage-inducible protein2 (chu_0679) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
|
|
|
| 37 | c2i00D_ |
|
not modelled |
85.0 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:acetyltransferase, gnat family;
PDBTitle: crystal structure of acetyltransferase (gnat family) from enterococcus2 faecalis
|
|
|
| 38 | c4nssA_ |
|
not modelled |
82.6 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:mycobacterial protein;
PDBTitle: a structural and functional investigation of a novel protein from2 mycobacterium smegmatis implicated in mycobacterial macrophage3 survivability
|
|
|
| 39 | d2nsfa2 |
|
not modelled |
80.6 |
14 |
Fold:SCP-like
Superfamily:SCP-like
Family:Micothiol-dependent maleylpyruvate isomerase C-terminal domain-like |
|
|
| 40 | c2ozgA_ |
|
not modelled |
58.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:gcn5-related n-acetyltransferase;
PDBTitle: crystal structure of gcn5-related n-acetyltransferase (yp_325469.1)2 from anabaena variabilis atcc 29413 at 2.00 a resolution
|
|
|
| 41 | d2i00a1 |
|
not modelled |
51.8 |
12 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
|
|
| 42 | d2ozga1 |
|
not modelled |
50.4 |
14 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
|
|
| 43 | d2hv2a1 |
|
not modelled |
42.4 |
11 |
Fold:SCP-like
Superfamily:SCP-like
Family:EF1021 C-terminal domain-like |
|
|
| 44 | c6qkjA_ |
|
not modelled |
41.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: egtb from chloracidobacterium thermophilum, a type ii sulfoxide2 synthase in complex with n,n,n-trimethyl-histidine
|
|
|
| 45 | d1wfra_ |
|
not modelled |
38.8 |
24 |
Fold:SCP-like
Superfamily:SCP-like
Family:Sterol carrier protein, SCP |
|
|
| 46 | d1w2za3 |
|
not modelled |
32.0 |
15 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
|
|
| 47 | c2dkzA_ |
|
not modelled |
29.6 |
22 |
PDB header:signaling protein
Chain: A: PDB Molecule:hypothetical protein loc64762;
PDBTitle: solution structure of the sam_pnt-domain of the2 hypothetical protein loc64762
|
|
|
| 48 | c2jobA_ |
|
not modelled |
25.0 |
60 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:antilipopolysaccharide factor;
PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
|
|
|
| 49 | c2hv2D_ |
|
not modelled |
24.6 |
19 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of conserved protein of unknown function from2 enterococcus faecalis v583 at 2.4 a resolution, probable n-3 acyltransferase
|
|
|
| 50 | c3n7zD_ |
|
not modelled |
22.7 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:acetyltransferase, gnat family;
PDBTitle: crystal structure of acetyltransferase from bacillus anthracis
|
|
|
| 51 | c4my3A_ |
|
not modelled |
14.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:gcn5-related n-acetyltransferase;
PDBTitle: crystal structure of gcn5-related n-acetyltransferase from kribbella2 flavida
|
|
|
| 52 | c6rftB_ |
|
not modelled |
11.3 |
14 |
PDB header:antibiotic
Chain: B: PDB Molecule:uncharacterized n-acetyltransferase d2e36_21790;
PDBTitle: crystal structure of eis2 from mycobacterium abscessus bound to2 acetyl-coa
|
|
|
| 53 | c3sxnC_ |
|
not modelled |
10.9 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:enhanced intracellular survival protein;
PDBTitle: mycobacterium tuberculosis eis protein initiates modulation of host2 immune responses by acetylation of dusp16/mkp-7
|
|
|
| 54 | c3r1kA_ |
|
not modelled |
9.3 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:enhanced intracellular survival protein;
PDBTitle: crystal structure of acetyltransferase eis from mycobacterium2 tuberculosis h37rv in complex with coa and an acetamide moiety
|
|
|
| 55 | c4idiA_ |
|
not modelled |
9.2 |
6 |
PDB header:protein binding
Chain: A: PDB Molecule:oryza sativa rurm1-related;
PDBTitle: crystal structure of rurm1-related protein from plasmodium yoelii,2 py06420
|
|
|
| 56 | d1t8sa_ |
|
not modelled |
7.3 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
|
|
| 57 | d1iiea_ |
|
not modelled |
6.0 |
40 |
Fold:Class II MHC-associated invariant chain ectoplasmic trimerization domain
Superfamily:Class II MHC-associated invariant chain ectoplasmic trimerization domain
Family:Class II MHC-associated invariant chain ectoplasmic trimerization domain |
|
|
| 58 | c2iv1J_ |
|
not modelled |
5.9 |
6 |
PDB header:lyase
Chain: J: PDB Molecule:cyanate hydratase;
PDBTitle: site directed mutagenesis of key residues involved in the catalytic2 mechanism of cyanase
|
|
|
| 59 | d1lbaa_ |
|
not modelled |
5.7 |
27 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
|
|