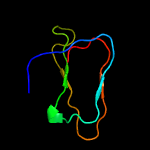
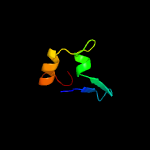
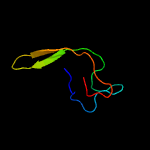
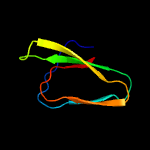
1 c5zeyC_
100.0
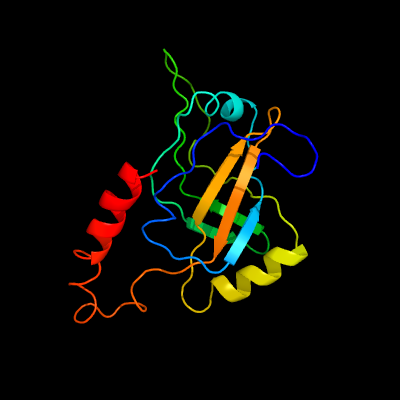
76
PDB header: ribosomeChain: C: PDB Molecule: ssra-binding protein;PDBTitle: m. smegmatis trans-translation state 70s ribosome
2 d1p6va_
100.0
50
Fold: Small protein B (SmpB)Superfamily: Small protein B (SmpB)Family: Small protein B (SmpB)
3 c1p6vC_
100.0
50
PDB header: rna binding protein/rnaChain: C: PDB Molecule: ssra-binding protein;PDBTitle: crystal structure of the trna domain of transfer-messenger2 rna in complex with smpb
4 c1j1hA_
100.0
52
PDB header: rna binding proteinChain: A: PDB Molecule: small protein b;PDBTitle: solution structure of a tmrna-binding protein, smpb, from2 thermus thermophilus
5 d1wjxa_
100.0
53
Fold: Small protein B (SmpB)Superfamily: Small protein B (SmpB)Family: Small protein B (SmpB)
6 c6hbeA_
36.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: copper-containing nitrite reductase;PDBTitle: cu-containing nitrite reductase (nirk) from thermus scotoductus sa-01
7 d2ipqx1
31.9
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: STY4665 C-terminal domain-like
8 c2qnkA_
21.3
30
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxyanthranilate 3,4-dioxygenase;PDBTitle: crystal structure of human 3-hydroxyanthranilate 3,4-dioxygenase
9 d1sdwa2
19.0
15
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: PHM/PNGase FFamily: Peptidylglycine alpha-hydroxylating monooxygenase, PHM
10 c4q28B_
15.9
23
PDB header: structural proteinChain: B: PDB Molecule: periplakin;PDBTitle: crystal structure of the plectin 1 and 2 repeats of the human2 periplakin. northeast structural genomics consortium (nesg) target3 hr9083a
11 c1wqsA_
15.5
14
PDB header: hydrolaseChain: A: PDB Molecule: 3c-like protease;PDBTitle: crystal structure of norovirus 3c-like protease
12 d1g31a_
15.3
27
Fold: GroES-likeSuperfamily: GroES-likeFamily: GroES
13 d1yfua1
14.3
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like
14 c4u39L_
13.9
45
PDB header: cell cycleChain: L: PDB Molecule: cell division factor;PDBTitle: crystal structure of ftsz:mciz complex from bacillus subtilis
15 c6b6iD_
13.4
18
PDB header: viral protein,proteaseChain: D: PDB Molecule: 3c-like protease;PDBTitle: 2.4a resolution structure of human norovirus gii.4 protease
16 c4u39J_
13.2
45
PDB header: cell cycleChain: J: PDB Molecule: cell division factor;PDBTitle: crystal structure of ftsz:mciz complex from bacillus subtilis
17 c6pqhA_
12.8
29
PDB header: ligaseChain: A: PDB Molecule: asparagine--trna ligase;PDBTitle: crystal structure of asparagine-trna ligase from elizabethkingia sp.2 ccug 26117
18 c2elvA_
12.1
38
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 6th c2h2 zinc finger of human2 zinc finger protein 406
19 c4ij2E_
11.1
23
PDB header: oxygen transport/protein bindingChain: E: PDB Molecule: iron-regulated surface determinant protein h;PDBTitle: human methemoglobin in complex with the second and third neat domains2 of isdh from staphylococcus aureus
20 c3jsbA_
11.0
17
PDB header: rna binding proteinChain: A: PDB Molecule: rna-directed rna polymerase;PDBTitle: crystal structure of the n-terminal domain of the lymphocytic2 choriomeningitis virus l protein
21 c5t2tB_
not modelled
10.8
17
PDB header: transferaseChain: B: PDB Molecule: rna-directed rna polymerase l;PDBTitle: crystal structure of lymphocytic choriomeningitis mammarenavirus2 endonuclease bound to compound l742001
22 c1hf9B_
not modelled
10.2
29
PDB header: atpase inhibitorChain: B: PDB Molecule: atpase inhibitor (mitochondrial);PDBTitle: c-terminal coiled-coil domain from bovine if1
23 c4i1tA_
not modelled
10.2
28
PDB header: viral proteinChain: A: PDB Molecule: rna-directed rna polymerase l;PDBTitle: crystal structure of the cap-snatching endonuclease from pichinde2 virus
24 c6nwoD_
not modelled
9.7
12
PDB header: transcriptionChain: D: PDB Molecule: transcriptional regulator bgar;PDBTitle: structures of the transcriptional regulator bgar, a lactose sensor.
25 d1zvfa1
not modelled
9.6
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like
26 c2quoA_
not modelled
9.3
27
PDB header: toxinChain: A: PDB Molecule: heat-labile enterotoxin b chain;PDBTitle: crystal structure of c terminal fragment of clostridium perfringens2 enterotoxin
27 c5hgqD_
not modelled
9.3
30
PDB header: ligase/ligase inhibitorChain: D: PDB Molecule: lysine--trna ligase;PDBTitle: loa loa lysyl-trna synthetase in complex with cladosporin.
28 c4u39N_
not modelled
9.1
36
PDB header: cell cycleChain: N: PDB Molecule: cell division factor;PDBTitle: crystal structure of ftsz:mciz complex from bacillus subtilis
29 d1d1na_
not modelled
8.8
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors
30 c5xumA_
not modelled
8.8
22
PDB header: transferaseChain: A: PDB Molecule: holo-[acyl-carrier-protein] synthase;PDBTitle: crystal structure of thermotoga maritima holo-[acyl-carrier-protein]2 synthase (acps)
31 c2oa9B_
not modelled
8.7
24
PDB header: hydrolaseChain: B: PDB Molecule: r.mvai;PDBTitle: restriction endonuclease mvai in the absence of dna
32 c4miwA_
not modelled
8.6
22
PDB header: viral protein, transferaseChain: A: PDB Molecule: rna-directed rna polymerase l;PDBTitle: high-resolution structure of the n-terminal endonuclease domain of the2 lassa virus l polymerase
33 c3mjdA_
not modelled
8.5
31
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.9 angstrom crystal structure of orotate phosphoribosyltransferase2 (pyre) francisella tularensis.
34 c2w3zA_
not modelled
8.0
12
PDB header: hydrolaseChain: A: PDB Molecule: putative deacetylase;PDBTitle: structure of a streptococcus mutans ce4 esterase
35 c4tq0D_
not modelled
7.6
29
PDB header: protein bindingChain: D: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg5-atg16n69
36 c4tq0B_
not modelled
7.5
29
PDB header: protein bindingChain: B: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg5-atg16n69
37 d2c0ha1
not modelled
7.3
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
38 d2cc0a1
not modelled
7.3
15
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
39 d2c1ia1
not modelled
7.1
16
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase
40 d2nlya1
not modelled
7.1
23
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: Divergent polysaccharide deacetylase
41 c4l1gB_
not modelled
6.6
24
PDB header: hydrolaseChain: B: PDB Molecule: peptidoglycan n-acetylglucosamine deacetylase;PDBTitle: crystal structure of the bc1960 peptidoglycan n-acetylglucosamine2 deacetylase from bacillus cereus
42 c5npvB_
not modelled
6.6
29
PDB header: cell cycleChain: B: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (i4)
43 c4m1bA_
not modelled
6.5
20
PDB header: hydrolaseChain: A: PDB Molecule: polysaccharide deacetylase;PDBTitle: structural determination of ba0150, a polysaccharide deacetylase from2 bacillus anthracis
44 c5npvD_
not modelled
6.4
29
PDB header: cell cycleChain: D: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (i4)
45 c5npwF_
not modelled
6.4
29
PDB header: cell cycleChain: F: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (c2)
46 c3rpmA_
not modelled
6.4
23
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetyl-hexosaminidase;PDBTitle: crystal structure of the first gh20 domain of a novel beta-n-acetyl-2 hexosaminidase strh from streptococcus pneumoniae r6
47 c2ylaA_
not modelled
6.3
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-n-acetylhexosaminidase;PDBTitle: inhibition of the pneumococcal virulence factor strh and2 molecular insights into n-glycan recognition and3 hydrolysis
48 c4u1eB_
not modelled
6.3
35
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: crystal structure of the eif3b-ctd/eif3i/eif3g-ntd translation2 initiation complex
49 c5jp6A_
not modelled
6.3
11
PDB header: hydrolaseChain: A: PDB Molecule: putative polysaccharide deacetylase;PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
50 d1im3d_
not modelled
6.2
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytomegalovirus protein US2
51 d1h2ca_
not modelled
6.1
9
Fold: EV matrix proteinSuperfamily: EV matrix proteinFamily: EV matrix protein
52 d1kk1a1
not modelled
6.1
32
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors
53 d8ohma2
not modelled
6.1
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RNA helicase
54 c2r2yA_
not modelled
6.1
29
PDB header: protein bindingChain: A: PDB Molecule: protein adrm1;PDBTitle: crystal structure of the proteasomal rpn13 pru-domain
55 c2z59A_
not modelled
6.1
29
PDB header: protein transportChain: A: PDB Molecule: protein adrm1;PDBTitle: complex structures of mouse rpn13 (22-130aa) and ubiquitin
56 c5x8r8_
not modelled
6.1
25
PDB header: ribosomeChain: 8: PDB Molecule: 30s ribosomal protein s1, chloroplastic;PDBTitle: structure of the 30s small subunit of chloroplast ribosome from2 spinach
57 c1h2dA_
not modelled
6.1
9
PDB header: virus/viral proteinChain: A: PDB Molecule: matrix protein vp40;PDBTitle: ebola virus matrix protein vp40 n-terminal domain in complex with rna2 (low-resolution vp40[31-212] variant).
58 d2ieca1
not modelled
5.9
71
Fold: MK0786-likeSuperfamily: MK0786-likeFamily: MK0786-like
59 c1gmjD_
not modelled
5.9
29
PDB header: atpase inhibitorChain: D: PDB Molecule: atpase inhibitor;PDBTitle: the structure of bovine if1, the regulatory subunit of mitochondrial2 f-atpase
60 d2i52a1
not modelled
5.6
71
Fold: MK0786-likeSuperfamily: MK0786-likeFamily: MK0786-like
61 c4uecB_
not modelled
5.6
29
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 4g, isoform a;PDBTitle: complex of d. melanogaster eif4e with eif4g and cap analog
62 c4h3wB_
not modelled
5.5
47
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative secreted protein (bdi_1231) from2 parabacteroides distasonis atcc 8503 at 2.00 a resolution
63 c4ah6B_
not modelled
5.5
24
PDB header: ligaseChain: B: PDB Molecule: aspartate--trna ligase, mitochondrial;PDBTitle: human mitochondrial aspartyl-trna synthetase
64 d1jaka1
not modelled
5.3
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain
65 c2ogfD_
not modelled
5.2
57
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein mj0408;PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
66 d1yhta1
not modelled
5.2
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain
67 c3wzhA_
not modelled
5.2
18
PDB header: transcriptionChain: A: PDB Molecule: uncharacterized protein af_1864;PDBTitle: crystal structure of afcsx3
68 d1mkya3
not modelled
5.2
25
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Probable GTPase Der, C-terminal domainFamily: Probable GTPase Der, C-terminal domain
69 d2qfaa1
not modelled
5.2
23
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat
70 d2gjxa1
not modelled
5.1
5
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain