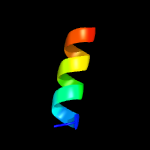| 1 |
|
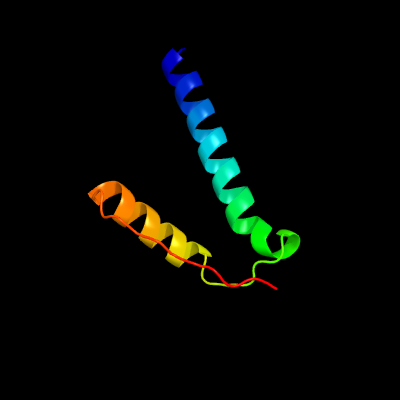
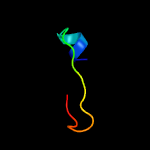
PDB 2fb5 chain A domain 1
Region: 4 - 61
Aligned: 56
Modelled: 58
Confidence: 48.2%
Identity: 21%
Fold: YojJ-like
Superfamily: YojJ-like
Family: YojJ-like
Phyre2
| 2 |
|
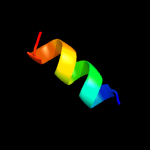
PDB 3brv chain B
Region: 23 - 46
Aligned: 24
Modelled: 24
Confidence: 23.2%
Identity: 38%
PDB header:transferase/transcription
Chain: B: PDB Molecule:nf-kappa-b essential modulator;
PDBTitle: nemo/ikkb association domain structure
Phyre2
| 3 |
|
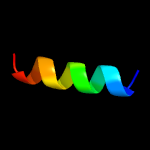
PDB 3msv chain B
Region: 73 - 91
Aligned: 19
Modelled: 19
Confidence: 14.5%
Identity: 16%
PDB header:protein binding
Chain: B: PDB Molecule:nuclear import adaptor, nro1;
PDBTitle: the hypoxic regulator of sterol synthesis nro1 is a nuclear import2 adaptor
Phyre2
| 4 |
|
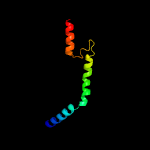
PDB 1yod chain B
Region: 16 - 29
Aligned: 14
Modelled: 14
Confidence: 10.8%
Identity: 50%
PDB header:de novo protein
Chain: B: PDB Molecule:water-solublized phospholamban;
PDBTitle: crystal structure of a water soluble analog of phospholamban
Phyre2
| 5 |
|
PDB 1yod chain A
Region: 16 - 29
Aligned: 14
Modelled: 14
Confidence: 9.9%
Identity: 50%
PDB header:de novo protein
Chain: A: PDB Molecule:water-solublized phospholamban;
PDBTitle: crystal structure of a water soluble analog of phospholamban
Phyre2
| 6 |
|
PDB 3qtm chain B
Region: 80 - 91
Aligned: 12
Modelled: 12
Confidence: 9.9%
Identity: 25%
PDB header:translation
Chain: B: PDB Molecule:uncharacterized protein c4b3.07;
PDBTitle: structure of s. pombe nuclear import adaptor nro1 (space group p21)
Phyre2
| 7 |
|
PDB 3jqh chain A
Region: 33 - 47
Aligned: 15
Modelled: 15
Confidence: 9.5%
Identity: 53%
PDB header:sugar binding protein
Chain: A: PDB Molecule:c-type lectin domain family 4 member m;
PDBTitle: structure of the neck region of the glycan-binding receptor dc-signr
Phyre2
| 8 |
|
PDB 5iy3 chain A
Region: 88 - 96
Aligned: 9
Modelled: 9
Confidence: 9.1%
Identity: 44%
PDB header:viral protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: zika virus non-structural protein ns1
Phyre2
| 9 |
|
PDB 5yxa chain C
Region: 87 - 96
Aligned: 10
Modelled: 10
Confidence: 8.7%
Identity: 30%
PDB header:viral protein
Chain: C: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of the c-terminal fragment of ns1 protein from2 yellow fever virus
Phyre2
| 10 |
|
PDB 6rdf chain 7
Region: 27 - 90
Aligned: 58
Modelled: 64
Confidence: 8.7%
Identity: 26%
PDB header:proton transport
Chain: 7: PDB Molecule:mitochondrial atp synthase associated protein asa7;
PDBTitle: cryoem structure of polytomella f-atp synthase, primary rotary state2 3, monomer-masked refinement
Phyre2
| 11 |
|
PDB 4o6c chain B
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 8.1%
Identity: 45%
PDB header:viral protein
Chain: B: PDB Molecule:ns1;
PDBTitle: west nile virus non-structural protein 1 (ns1) form 2 crystal
Phyre2
| 12 |
|
PDB 1zps chain A domain 1
Region: 61 - 79
Aligned: 19
Modelled: 19
Confidence: 8.0%
Identity: 37%
Fold: HisI-like
Superfamily: HisI-like
Family: HisI-like
Phyre2
| 13 |
|
PDB 5k6k chain B
Region: 88 - 96
Aligned: 9
Modelled: 9
Confidence: 7.5%
Identity: 44%
PDB header:viral protein
Chain: B: PDB Molecule:zika virus protein;
PDBTitle: zika virus non-structural protein 1 (ns1)
Phyre2
| 14 |
|
PDB 2db7 chain A domain 1
Region: 20 - 41
Aligned: 22
Modelled: 22
Confidence: 7.3%
Identity: 36%
Fold: Orange domain-like
Superfamily: Orange domain-like
Family: Hairy Orange domain
Phyre2
| 15 |
|
PDB 2lql chain A
Region: 23 - 32
Aligned: 10
Modelled: 10
Confidence: 7.3%
Identity: 50%
PDB header:protein binding
Chain: A: PDB Molecule:coiled-coil-helix-coiled-coil-helix domain-containing
PDBTitle: solution structure of chch5
Phyre2
| 16 |
|
PDB 1v54 chain I
Region: 3 - 45
Aligned: 43
Modelled: 43
Confidence: 6.2%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIc
Family: Mitochondrial cytochrome c oxidase subunit VIc
Phyre2
| 17 |
|
PDB 1dnp chain A domain 1
Region: 35 - 88
Aligned: 41
Modelled: 42
Confidence: 6.0%
Identity: 27%
Fold: Cryptochrome/photolyase FAD-binding domain
Superfamily: Cryptochrome/photolyase FAD-binding domain
Family: Cryptochrome/photolyase FAD-binding domain
Phyre2
| 18 |
|
PDB 4o6b chain A
Region: 88 - 96
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 56%
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: dengue type2 virus non-structural protein 1 (ns1) form 1 crystal
Phyre2
| 19 |
|
PDB 2n2u chain A
Region: 92 - 96
Aligned: 5
Modelled: 5
Confidence: 5.8%
Identity: 100%
PDB header:unknown function, structural genomics
Chain: A: PDB Molecule:or358;
PDBTitle: solution nmr structure of de novo designed ferredoxin fold protein2 sfr3, northeast structural genomics consortium (nesg) target or358
Phyre2
| 20 |
|
PDB 5yyl chain C
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 5.6%
Identity: 56%
PDB header:signaling protein
Chain: C: PDB Molecule:apisimin;
PDBTitle: structure of major royal jelly protein 1 oligomer
Phyre2
| 21 |
|