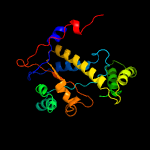
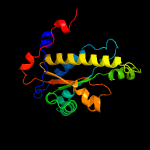
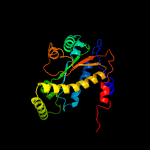
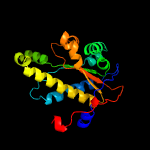
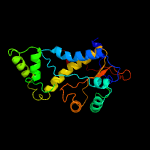
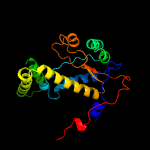
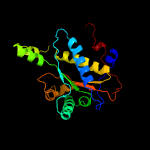
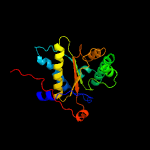
1 c2ymvA_
100.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: acg nitroreductase;PDBTitle: structure of reduced m smegmatis 5246, a homologue of m.2 tuberculosis acg
2 c3gr3B_
100.0
16
PDB header: flavoproteinChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a nitroreductase-like family protein (pnba,2 bh06130) from bartonella henselae str. houston-1 at 1.45 a resolution
3 c2wzvB_
100.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: nfnb protein;PDBTitle: crystal structure of the fmn-dependent nitroreductase nfnb2 from mycobacterium smegmatis
4 c3gh8A_
100.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: iodotyrosine dehalogenase 1;PDBTitle: crystal structure of mus musculus iodotyrosine deiodinase (iyd) bound2 to fmn and di-iodotyrosine (dit)
5 c2islB_
100.0
16
PDB header: flavoproteinChain: B: PDB Molecule: blub;PDBTitle: blub bound to reduced flavin (fmnh2) and molecular oxygen.2 (clear crystal form)
6 c4eo3A_
100.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: bacterioferritin comigratory protein/nadh dehydrogenase;PDBTitle: peroxiredoxin nitroreductase fusion enzyme
7 c5ko8B_
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of haliscomenobacter hydrossis iodotyrosine2 deiodinase (iyd) bound to fmn and mono-iodotyrosine (i-tyr)
8 c3eo8A_
99.9
14
PDB header: flavoproteinChain: A: PDB Molecule: blub-like flavoprotein;PDBTitle: crystal structure of blub-like flavoprotein (yp_001089088.1) from2 clostridium difficile 630 at 1.74 a resolution
9 d1vfra_
99.9
11
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
10 d1zcha1
99.9
14
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
11 d1noxa_
99.9
21
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
12 c5hdjA_
99.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: nfra1;PDBTitle: structure of b. megaterium nfra1
13 c3k6hB_
99.9
20
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a nitroreductase family protein from2 agrobacterium tumefaciens str. c58
14 c5heiE_
99.9
19
PDB header: oxidoreductaseChain: E: PDB Molecule: nfra2;PDBTitle: structure of b. megaterium nfra2
15 d1bkja_
99.9
22
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
16 c3bemA_
99.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nad(p)h nitroreductase ydfn;PDBTitle: crystal structure of putative nitroreductase ydfn (2632848) from2 bacillus subtilis at 1.65 a resolution
17 c2hayD_
99.9
15
PDB header: oxidoreductaseChain: D: PDB Molecule: putative nad(p)h-flavin oxidoreductase;PDBTitle: the crystal structure of the putative nad(p)h-flavin oxidoreductase2 from streptococcus pyogenes m1 gas
18 c6czpH_
99.9
12
PDB header: oxidoreductaseChain: H: PDB Molecule: oxygen-insensitive nad(p)h nitroreductase;PDBTitle: 2.2 angstrom resolution crystal structure oxygen-insensitive nad(p)h-2 dependent nitroreductase nfsb from vibrio vulnificus in complex with3 fmn
19 c3n2sD_
99.9
19
PDB header: oxidoreductaseChain: D: PDB Molecule: nadph-dependent nitro/flavin reductase;PDBTitle: structure of nfra1 nitroreductase from b. subtilis
20 c3ek3A_
99.9
15
PDB header: flavoproteinChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of nitroreductase with bound fmn (yp_211706.1) from2 bacteroides fragilis nctc 9343 at 1.70 a resolution
21 d1ykia1
not modelled
99.9
12
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
22 c2i7hE_
not modelled
99.9
13
PDB header: oxidoreductaseChain: E: PDB Molecule: nitroreductase-like family protein;PDBTitle: crystal structure of the nitroreductase-like family protein from2 bacillus cereus
23 c3eofB_
not modelled
99.9
17
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of putative oxidoreductase (yp_213212.1) from2 bacteroides fragilis nctc 9343 at 1.99 a resolution
24 c3gfaB_
not modelled
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (cd3205) from clostridium difficile 630 at 1.35 a resolution
25 c3ge6B_
not modelled
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (exig_2970) from exiguobacterium sibiricum 255-15 at 1.85 a3 resolution
26 d1f5va_
not modelled
99.9
24
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
27 c4xomB_
not modelled
99.9
17
PDB header: unknown functionChain: B: PDB Molecule: coenzyme f420:l-glutamate ligase;PDBTitle: coenzyme f420:l-glutamate ligase (fbib) from mycobacterium2 tuberculosis (c-terminal domain).
28 c3ge5A_
not modelled
99.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nad(p)h:fmn oxidoreductase;PDBTitle: crystal structure of a putative nad(p)h:fmn oxidoreductase (pg0310)2 from porphyromonas gingivalis w83 at 1.70 a resolution
29 c3gbhC_
not modelled
99.9
12
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p)h-flavin oxidoreductase;PDBTitle: crystal structure of a putative nad(p)h:fmn oxidoreductase (se1966)2 from staphylococcus epidermidis atcc 12228 at 2.00 a resolution
30 c3e39A_
not modelled
99.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase in complex with fmn2 (dde_0787) from desulfovibrio desulfuricans subsp. at 1.70 a3 resolution
31 c3to0A_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: iodotyrosine deiodinase 1;PDBTitle: crystal structure of mus musculus iodotyrosine deiodinase (iyd) c217a,2 c239a bound to fmn
32 c4qlyB_
not modelled
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: enone reductase cla-er;PDBTitle: crystal structure of cla-er, a novel enone reductase catalyzing a key2 step of a gut-bacterial fatty acid saturation metabolism,3 biohydrogenation
33 c3gagB_
not modelled
99.9
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadh dehydrogenase, nadph nitroreductase;PDBTitle: crystal structure of a nitroreductase-like protein (smu.346) from2 streptococcus mutans at 1.70 a resolution
34 c3of4A_
not modelled
99.9
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a fmn/fad- and nad(p)h-dependent nitroreductase2 (nfnb, il2077) from idiomarina loihiensis l2tr at 1.90 a resolution
35 d2b67a1
not modelled
99.9
14
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
36 c3pxvD_
not modelled
99.9
15
PDB header: oxidoreductaseChain: D: PDB Molecule: nitroreductase;PDBTitle: crystal structure of a nitroreductase with bound fmn (dhaf_2018) from2 desulfitobacterium hafniense dcb-2 at 2.30 a resolution
37 d1kqba_
not modelled
99.9
14
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
38 c3kwkA_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh dehydrogenase/nad(p)h nitroreductase;PDBTitle: crystal structure of putative nadh dehydrogenase/nad(p)h2 nitroreductase (np_809094.1) from bacteroides thetaiotaomicron vpi-3 5482 at 1.54 a resolution
39 d1ywqa1
not modelled
99.9
11
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
40 c3koqC_
not modelled
99.9
19
PDB header: oxidoreductaseChain: C: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a nitroreductase family protein (cd3355) from2 clostridium difficile 630 at 1.58 a resolution
41 d2ifaa1
not modelled
99.9
11
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
42 c2h0uA_
not modelled
99.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph-flavin oxidoreductase;PDBTitle: crystal structure of nad(p)h-flavin oxidoreductase from helicobacter2 pylori
43 c2r01A_
not modelled
99.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of a putative fmn-dependent nitroreductase (ct0345)2 from chlorobium tepidum tls at 1.15 a resolution
44 c3m5kA_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh dehydrogenase/nad(p)h nitroreductase;PDBTitle: crystal structure of putative nadh dehydrogenase/nad(p)h2 nitroreductase (bdi_1728) from parabacteroides distasonis atcc 85033 at 1.86 a resolution
45 c4dn2A_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nitroreductase;PDBTitle: crystal structure of putative nitroreductase from geobacter2 metallireducens gs-15
46 d2frea1
not modelled
99.9
14
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: NADH oxidase/flavin reductase
47 c3qdlD_
not modelled
99.9
13
PDB header: oxidoreductaseChain: D: PDB Molecule: oxygen-insensitive nadph nitroreductase;PDBTitle: crystal structure of rdxa from helicobacter pyroli
48 c3e10B_
not modelled
99.9
16
PDB header: oxidoreductaseChain: B: PDB Molecule: putative nadh oxidase;PDBTitle: crystal structure of putative nadh oxidase (np_348178.1) from2 clostridium acetobutylicum at 1.40 a resolution
49 c3g14B_
not modelled
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nitroreductase family protein;PDBTitle: crystal structure of nitroreductase family protein (yp_877874.1) from2 clostridium novyi nt at 1.75 a resolution
50 c2wqfA_
not modelled
99.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: copper induced nitroreductase d;PDBTitle: crystal structure of the nitroreductase cind from2 lactococcus lactis in complex with fmn
51 c5j6cA_
not modelled
99.9
20
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase;PDBTitle: fmn-dependent nitroreductase (cdr20291_0767) from clostridium2 difficile r20291
52 c3bm2B_
not modelled
99.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: protein ydja;PDBTitle: crystal structure of a minimal nitroreductase ydja from escherichia2 coli k12 with and without fmn cofactor
53 c5j62B_
not modelled
99.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: putative reductase;PDBTitle: fmn-dependent nitroreductase (cdr20291_0684) from clostridium2 difficile r20291
54 c3hj9A_
not modelled
99.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of a putative nitroreductase (reut_a1228) from2 ralstonia eutropha jmp134 at 2.00 a resolution
55 c3hoiA_
not modelled
99.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nitroreductase bf3017;PDBTitle: crystal structure of fmn-dependent nitroreductase bf3017 from2 bacteroides fragilis nctc 9343 (yp_212631.1) from bacteroides3 fragilis nctc 9343 at 1.55 a resolution
56 c4urpB_
not modelled
99.8
14
PDB header: oxidoreductaseChain: B: PDB Molecule: fatty acid repression mutant protein 2;PDBTitle: the crystal structure of nitroreductase from saccharomyces2 cerevisiae
57 d1vkwa_
not modelled
99.7
21
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: Putative nitroreductase TM1586
58 c3eo7A_
not modelled
99.7
20
PDB header: flavoproteinChain: A: PDB Molecule: putative nitroreductase;PDBTitle: crystal structure of a putative nitroreductase (ava_2154) from2 anabaena variabilis atcc 29413 at 1.80 a resolution
59 c5lq4B_
not modelled
99.7
21
PDB header: oxidoreductaseChain: B: PDB Molecule: cyagox;PDBTitle: the structure of thcox, the first oxidase protein from the cyanobactin2 pathways
60 c6gosC_
not modelled
98.6
13
PDB header: antibiotic/inhibitorChain: C: PDB Molecule: microcin b17-processing protein mcbc;PDBTitle: e. coli microcin synthetase mcbbcd complex with pro-mccb17 bound
61 c3vteA_
not modelled
55.4
7
PDB header: oxidoreductaseChain: A: PDB Molecule: tetrahydrocannabinolic acid synthase;PDBTitle: crystal structure of tetrahydrocannabinolic acid synthase from2 cannabis sativa
62 d1f3ub_
not modelled
24.8
38
Fold: triple barrelSuperfamily: Rap30/74 interaction domainsFamily: Rap30/74 interaction domains
63 c4kdiC_
not modelled
21.2
38
PDB header: signaling protein/hydrolaseChain: C: PDB Molecule: ubiquitin thioesterase otu1;PDBTitle: crystal structure of p97/vcp n in complex with otu1 ubxl
64 c3fwaA_
not modelled
18.5
9
PDB header: flavoproteinChain: A: PDB Molecule: reticuline oxidase;PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
65 c5sxpF_
not modelled
18.3
50
PDB header: signaling protein/ligaseChain: F: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
66 d2pnwa1
not modelled
15.8
13
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: MLTA-like
67 c5sxpG_
not modelled
11.8
50
PDB header: signaling protein/ligaseChain: G: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
68 d2g5da1
not modelled
10.0
15
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: MLTA-like
69 c3d2hA_
not modelled
10.0
9
PDB header: oxidoreductaseChain: A: PDB Molecule: berberine bridge-forming enzyme;PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
70 d2qmsa1
not modelled
8.8
50
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain
71 d2fcia1
not modelled
8.5
40
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain
72 c3r74B_
not modelled
8.5
21
PDB header: lyase, biosynthetic proteinChain: B: PDB Molecule: anthranilate/para-aminobenzoate synthases component i;PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
73 c1lqlE_
not modelled
8.4
7
PDB header: unknown functionChain: E: PDB Molecule: osmotical inducible protein c like family;PDBTitle: crystal structure of osmc like protein from mycoplasma2 pneumoniae
74 d1lqla_
not modelled
8.4
7
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins
75 d1kjqa2
not modelled
8.1
7
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
76 c6hq9A_
not modelled
8.1
50
PDB header: hydrolaseChain: A: PDB Molecule: dna excision repair protein ercc-6-like 2;PDBTitle: crystal structure of the tudor domain of human ercc6-l2
77 c3fajA_
not modelled
7.9
35
PDB header: structural proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of the structural protein p131 of the archaeal virus2 acidianus two-tailed virus (atv)
78 d1ecfa2
not modelled
7.5
14
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Class II glutamine amidotransferases
79 d1lf6a2
not modelled
6.9
17
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Bacterial glucoamylase N-terminal domain-like
80 c2aapA_
not modelled
6.2
50
PDB header: toxinChain: A: PDB Molecule: jingzhaotoxin-vii;PDBTitle: solution structure of jingzhaotoxin-vii
81 d1kona_
not modelled
6.2
14
Fold: YebC-likeSuperfamily: YebC-likeFamily: YebC-like
82 c3tsjA_
not modelled
6.1
12
PDB header: allergen, oxidoreductaseChain: A: PDB Molecule: pollen allergen phl p 4;PDBTitle: crystal structure of phl p 4, a grass pollen allergen with glucose2 dehydrogenase activity
83 d2r4qa1
not modelled
5.8
25
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like
84 c4ml8C_
not modelled
5.8
5
PDB header: oxidoreductaseChain: C: PDB Molecule: cytokinin oxidase 2;PDBTitle: structure of maize cytokinin oxidase/dehydrogenase 2 (zmcko2)
85 d1ki0a1
not modelled
5.6
27
Fold: Kringle-likeSuperfamily: Kringle-likeFamily: Kringle modules
86 c2l2rA_
not modelled
5.4
57
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide ecamp1;PDBTitle: helical hairpin structure of a novel antimicrobial peptide ecamp1 from2 seeds of barnyard grass (echinochloa crus-galli)
87 c5abxB_
not modelled
5.3
18
PDB header: translationChain: B: PDB Molecule: 4e-binding protein mextli;PDBTitle: complex of c. elegans eif4e-3 with the 4e-binding protein2 mextli and cap analog
88 c2xu8B_
not modelled
5.3
33
PDB header: structural genomicsChain: B: PDB Molecule: pa1645;PDBTitle: structure of pa1645
89 d2bkwa1
not modelled
5.2
10
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like