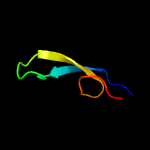
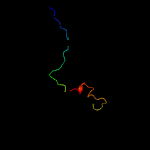
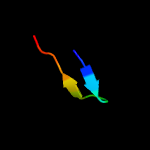
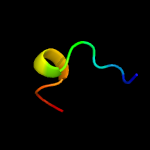
1 c6h9iB_
39.5
26
PDB header: rna binding proteinChain: B: PDB Molecule: csf5;PDBTitle: csf5, crispr-cas type iv cas6 crrna endonuclease
2 d1okga3
33.6
34
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: 3-mercaptopyruvate sulfurtransferase, C-terminal domain
3 c5v2sA_
16.6
17
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
4 c1nh1A_
15.1
26
PDB header: avirulence proteinChain: A: PDB Molecule: avirulence b protein;PDBTitle: crystal structure of the type iii effector avrb from2 pseudomonas syringae.
5 d1nh1a_
15.1
26
Fold: Antivirulence factorSuperfamily: Antivirulence factorFamily: Antivirulence factor
6 d1wvna1
14.9
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
7 c3sokB_
14.3
33
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
8 d1j4wa1
12.8
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
9 d1pama1
12.6
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
10 d2ctea1
11.9
37
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
11 c1okgA_
11.5
42
PDB header: transferaseChain: A: PDB Molecule: possible 3-mercaptopyruvate sulfurtransferase;PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
12 c4c47B_
11.5
41
PDB header: cell adhesionChain: B: PDB Molecule: inner membrane lipoprotein;PDBTitle: salmonella enterica trimeric lipoprotein sadb
13 c1gycA_
11.1
21
PDB header: oxidoreductaseChain: A: PDB Molecule: laccase 2;PDBTitle: crystal structure determination at room temperature of a laccase from2 trametes versicolor in its oxidised form containing a full complement3 of copper ions
14 d1el6a_
11.1
26
Fold: Baseplate structural protein gp11Superfamily: Baseplate structural protein gp11Family: Baseplate structural protein gp11
15 c2xlkB_
10.7
14
PDB header: hydrolase/rnaChain: B: PDB Molecule: csy4 endoribonuclease;PDBTitle: crystal structure of the csy4-crrna complex, orthorhombic form
16 c6j0mC_
10.5
20
PDB header: protein transportChain: C: PDB Molecule: pvc8;PDBTitle: cryo-em structure of an extracellular contractile injection system,2 pvc baseplate in extended state (reconstructed with c3 symmetry)
17 c2wj7D_
10.4
42
PDB header: chaperoneChain: D: PDB Molecule: alpha-crystallin b chain;PDBTitle: human alphab crystallin
18 d2ctda1
9.1
31
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
19 d1tj1a1
9.1
57
Fold: N-terminal domain of bifunctional PutA proteinSuperfamily: N-terminal domain of bifunctional PutA proteinFamily: N-terminal domain of bifunctional PutA protein
20 c6k4fU_
8.9
38
PDB header: biosynthetic proteinChain: U: PDB Molecule: duf1987 domain-containing protein;PDBTitle: siac of pseudomonas aeruginosa
21 c5craB_
not modelled
7.9
55
PDB header: hydrolaseChain: B: PDB Molecule: sdea;PDBTitle: structure of the sdea dub domain
22 d2pila_
not modelled
7.9
22
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin
23 c2hh3A_
not modelled
7.8
17
PDB header: rna binding proteinChain: A: PDB Molecule: kh-type splicing regulatory protein;PDBTitle: solution structure of the third kh domain of ksrp
24 d3bmva1
not modelled
7.7
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
25 c1v10A_
not modelled
7.6
28
PDB header: oxidaseChain: A: PDB Molecule: laccase;PDBTitle: structure of rigidoporus lignosus laccase from hemihedrally2 twinned crystals
26 d2ctma1
not modelled
7.5
31
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
27 c5dgoA_
not modelled
7.3
32
PDB header: cell cycleChain: A: PDB Molecule: cell division control protein 45 homolog;PDBTitle: crystal structure of cell division cycle protein 45 (cdc45)
28 c3kdpG_
not modelled
7.0
18
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
29 c3kdpH_
not modelled
7.0
18
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
30 c3lq9B_
not modelled
6.9
15
PDB header: signaling proteinChain: B: PDB Molecule: dna-damage-inducible transcript 4 protein;PDBTitle: crystal structure of human redd1, a hypoxia-induced regulator of mtor
31 d1x4na1
not modelled
6.9
11
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
32 d2nsfa2
not modelled
6.9
30
Fold: SCP-likeSuperfamily: SCP-likeFamily: Micothiol-dependent maleylpyruvate isomerase C-terminal domain-like
33 c3hj2B_
not modelled
6.8
27
PDB header: antimicrobial proteinChain: B: PDB Molecule: human neutrophil peptide 1;PDBTitle: crystal structure of covalent dimer of hnp1
34 c4hqjG_
not modelled
6.7
18
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
35 c2jp3A_
not modelled
6.6
29
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
36 c3qphA_
not modelled
6.5
14
PDB header: transcriptionChain: A: PDB Molecule: trmb, a global transcription regulator;PDBTitle: the three-dimensional structure of trmb, a global transcriptional2 regulator of the hyperthermophilic archaeon pyrococcus furiosus in3 complex with sucrose
37 c2m7gA_
not modelled
6.3
28
PDB header: cell adhesion, structural protein, electChain: A: PDB Molecule: geopilin domain 1 protein;PDBTitle: structure of the type iva major pilin from the electrically conductive2 bacterial nanowires of geobacter sulfurreducens
38 c6nbxG_
not modelled
6.3
20
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: t.elongatus ndh (data-set 2)
39 c5m73H_
not modelled
6.3
46
PDB header: rna binding proteinChain: H: PDB Molecule: signal recognition particle subunit srp72;PDBTitle: structure of the human srp s domain with srp72 rna-binding domain
40 d2axya1
not modelled
6.2
21
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
41 c4hqjE_
not modelled
6.1
18
PDB header: hydrolase/transport proteinChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
42 c2qdjA_
not modelled
6.1
18
PDB header: antitumor proteinChain: A: PDB Molecule: retinoblastoma-associated protein;PDBTitle: crystal structure of the retinoblastoma protein n-domain2 provides insight into tumor suppression, ligand3 interaction and holoprotein architecture
43 c6rbkC_
not modelled
6.1
22
PDB header: virus like particleChain: C: PDB Molecule: afp8;PDBTitle: cryo-em structure of the anti-feeding prophage (afp) baseplate in2 extended state, 3-fold symmetrised
44 c2wmmA_
not modelled
6.0
11
PDB header: cell cycleChain: A: PDB Molecule: chromosome partition protein mukb;PDBTitle: crystal structure of the hinge domain of mukb
45 c5b0uB_
not modelled
6.0
42
PDB header: luminescent proteinChain: B: PDB Molecule: oplophorus-luciferin 2-monooxygenase catalytic subunit;PDBTitle: crystal structure of the mutated 19 kda protein of oplophorus2 luciferase (nanokaz)
46 c2jo1A_
not modelled
5.8
24
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
47 c5wdaL_
not modelled
5.8
33
PDB header: protein transportChain: L: PDB Molecule: general secretion pathway protein g;PDBTitle: structure of the pulg pseudopilus
48 d1khma_
not modelled
5.7
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
49 d1oqwa_
not modelled
5.6
28
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin
50 c5m8bB_
not modelled
5.6
27
PDB header: hydrolaseChain: B: PDB Molecule: alpha-l-arabinofuranosidase ii;PDBTitle: crystal structure of alpha-l-arabinofuranosidase from lactobacillus2 brevis
51 d1zzka1
not modelled
5.6
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
52 c5xf9E_
not modelled
5.6
24
PDB header: oxidoreductaseChain: E: PDB Molecule: nad-reducing hydrogenase;PDBTitle: crystal structure of nad+-reducing [nife]-hydrogenase in the air-2 oxidized state
53 d1y7ba2
not modelled
5.5
22
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like
54 d1cgta1
not modelled
5.4
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
55 c2fugA_
not modelled
5.4
24
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase chain 1;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
56 c3uv0B_
not modelled
5.1
28
PDB header: protein bindingChain: B: PDB Molecule: mutator 2, isoform b;PDBTitle: crystal structure of the drosophila mu2 fha domain
57 d1cyga1
not modelled
5.1
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
58 c5lc5F_
not modelled
5.1
24
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh dehydrogenase [ubiquinone] flavoprotein 1,PDBTitle: structure of mammalian respiratory complex i, class2