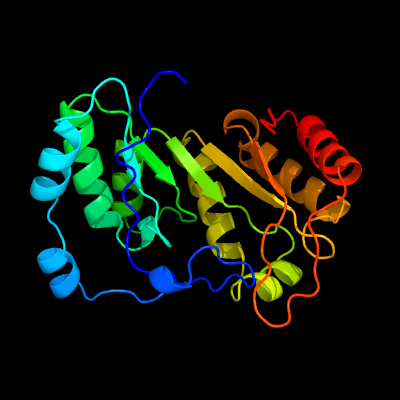
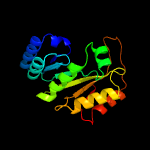
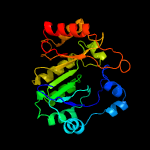
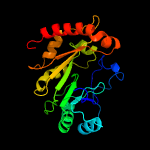
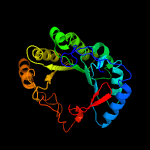
1 c3c8fA_
100.0
20
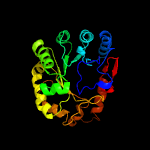
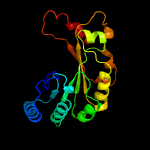
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with partially2 disordered adomet
2 c6fz6B_
99.9
17
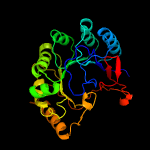
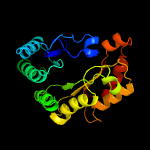
PDB header: transferaseChain: B: PDB Molecule: probable dual-specificity rna methyltransferase rlmn;PDBTitle: crystal structure of a radical sam methyltransferase from2 sphaerobacter thermophilus
3 c3rfaB_
99.9
13
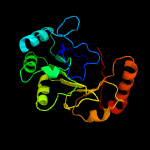
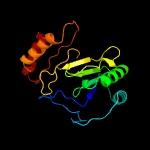
PDB header: oxidoreductaseChain: B: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
4 c3canA_
99.9
18
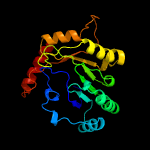
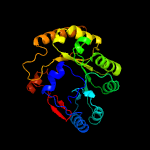
PDB header: lyase activatorChain: A: PDB Molecule: pyruvate-formate lyase-activating enzyme;PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
5 c3rfaA_
99.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
6 c2yx0A_
99.9
17
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam enzyme;PDBTitle: crystal structure of p. horikoshii tyw1
7 d1tv8a_
99.9
19
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: MoCo biosynthesis proteins
8 c5v1tA_
99.9
17
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam;PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
9 c6efnA_
99.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: sporulation killing factor maturation protein skfb;PDBTitle: structure of a ripp maturase, skfb
10 c4njkA_
99.9
21
PDB header: lyaseChain: A: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
11 c5th5C_
99.8
13
PDB header: lyaseChain: C: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
12 c6b4cH_
99.8
13
PDB header: antiviral proteinChain: H: PDB Molecule: viperin;PDBTitle: structure of viperin from trichoderma virens
13 c4u0pB_
99.8
16
PDB header: transferaseChain: B: PDB Molecule: lipoyl synthase 2;PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
14 c6nhlB_
99.8
15
PDB header: lyaseChain: B: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from escherichia coli
15 c4k39A_
99.8
16
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic sulfatase-maturating enzyme;PDBTitle: native ansmecpe with bound adomet and cp18cys peptide
16 c5vslB_
99.8
14
PDB header: antiviral proteinChain: B: PDB Molecule: radical s-adenosyl methionine domain-containing protein 2;PDBTitle: crystal structure of viperin with bound [4fe-4s] cluster and s-2 adenosylhomocysteine (sah)
17 c5wggA_
99.8
12
PDB header: peptide binding proteinChain: A: PDB Molecule: radical sam domain protein;PDBTitle: structural insights into thioether bond formation in the biosynthesis2 of sactipeptides
18 c4wcxC_
99.7
17
PDB header: lyaseChain: C: PDB Molecule: biotin and thiamin synthesis associated;PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
19 c5exkG_
99.7
16
PDB header: transferaseChain: G: PDB Molecule: lipoyl synthase;PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
20 c2z2uA_
99.7
21
PDB header: metal binding proteinChain: A: PDB Molecule: upf0026 protein mj0257;PDBTitle: crystal structure of archaeal tyw1
21 c2a5hC_
not modelled
99.6
13
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
22 d1r30a_
not modelled
99.6
17
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Biotin synthase
23 c1r30A_
not modelled
99.6
17
PDB header: transferaseChain: A: PDB Molecule: biotin synthase;PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
24 c3t7vA_
not modelled
99.6
13
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
25 c3cixA_
not modelled
99.6
13
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
26 c6c8vA_
not modelled
99.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: coenzyme pqq synthesis protein e;PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
27 c4jc0B_
not modelled
99.5
13
PDB header: transferaseChain: B: PDB Molecule: ribosomal protein s12 methylthiotransferase rimo;PDBTitle: crystal structure of thermotoga maritima holo rimo in complex with2 pentasulfide, northeast structural genomics consortium target vr77
28 c6fd2B_
not modelled
99.4
18
PDB header: biosynthetic proteinChain: B: PDB Molecule: putative apramycin biosynthetic oxidoreductase 4;PDBTitle: radical sam 1,2-diol dehydratase aprd4 in complex with its substrate2 paromamine
29 c4rtbA_
not modelled
99.3
12
PDB header: lyaseChain: A: PDB Molecule: hydg protein;PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
30 d1olta_
not modelled
99.3
11
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN
31 c4m7tA_
not modelled
99.2
14
PDB header: metal binding proteinChain: A: PDB Molecule: btrn;PDBTitle: crystal structure of btrn in complex with adomet and 2-doia
32 c5l7jA_
not modelled
99.2
15
PDB header: translationChain: A: PDB Molecule: elp3 family;PDBTitle: crystal structure of elp3 from dehalococcoides mccartyi
33 c5ul4A_
not modelled
99.0
11
PDB header: metal binding proteinChain: A: PDB Molecule: oxsb protein;PDBTitle: structure of cobalamin-dependent s-adenosylmethionine radical enzyme2 oxsb with aqua-cobalamin and s-adenosylmethionine bound
34 c4r33A_
not modelled
98.8
15
PDB header: lyaseChain: A: PDB Molecule: nosl;PDBTitle: x-ray structure of the tryptophan lyase nosl with tryptophan and s-2 adenosyl-l-homocysteine bound
35 c6qk7C_
not modelled
98.8
15
PDB header: translationChain: C: PDB Molecule: elongator complex protein 3;PDBTitle: elongator catalytic subcomplex elp123 lobe
36 c2qgqF_
not modelled
98.7
14
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein tm_1862;PDBTitle: crystal structure of tm_1862 from thermotoga maritima. northeast2 structural genomics consortium target vr77
37 c4fheA_
not modelled
98.7
11
PDB header: lyaseChain: A: PDB Molecule: spore photoproduct lyase;PDBTitle: spore photoproduct lyase c140a mutant
38 c6iazA_
not modelled
98.1
12
PDB header: transferaseChain: A: PDB Molecule: histone acetyltransferase, elp3 family;PDBTitle: the archaeal methanocaldococcus infernus elp3 with n-terminus deletion2 (1-46)
39 c3e49A_
not modelled
95.9
17
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849 with a tim barrel fold;PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
40 c3no5C_
not modelled
95.6
14
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
41 c2y0fD_
not modelled
95.1
27
PDB header: oxidoreductaseChain: D: PDB Molecule: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
42 c3c6cA_
not modelled
94.9
14
PDB header: hydrolaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
43 c3noyA_
not modelled
94.7
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;PDBTitle: crystal structure of ispg (gcpe)
44 c2y7eA_
not modelled
94.0
14
PDB header: lyaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
45 c3e02A_
not modelled
94.0
15
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849;PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
46 c3chvA_
not modelled
93.7
11
PDB header: metal binding proteinChain: A: PDB Molecule: prokaryotic domain of unknown function (duf849) with a timPDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
47 c5zmyF_
not modelled
92.9
15
PDB header: hydrolaseChain: F: PDB Molecule: cis-epoxysuccinate hydrolase;PDBTitle: crystal structure of a cis-epoxysuccinate hydrolase producing d(-)-2 tartaric acids
48 d1x7fa2
not modelled
92.8
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Outer surface protein, N-terminal domain
49 c1x7fA_
not modelled
92.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: outer surface protein;PDBTitle: crystal structure of an uncharacterized b. cereus protein
50 c4u3eA_
not modelled
88.0
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside triphosphate reductase;PDBTitle: anaerobic ribonucleotide reductase
51 c5fkvA_
not modelled
87.7
18
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii subunit alpha;PDBTitle: cryo-em structure of the e. coli replicative dna polymerase complex2 bound to dna (dna polymerase iii alpha, beta, epsilon, tau complex)
52 c2gb5B_
not modelled
86.2
16
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
53 c6o3pA_
not modelled
86.2
13
PDB header: hydrolaseChain: A: PDB Molecule: peroxisomal nadh pyrophosphatase nudt12;PDBTitle: crystal structure of the catalytic domain of mouse nudt12 in complex2 with amp and 3 mg2+ ions
54 c6hmsB_
not modelled
85.5
30
PDB header: replicationChain: B: PDB Molecule: dna polymerase ii large subunit,dna polymerase ii largePDBTitle: cryo-em map of dna polymerase d from pyrococcus abyssi in complex with2 dna
55 c5vrvA_
not modelled
83.4
18
PDB header: hydrolase,oxidoreductaseChain: A: PDB Molecule: protein regulated by acid ph;PDBTitle: 2.05 angstrom resolution crystal structure of c-terminal domain2 (duf2156) of putative lysylphosphatidylglycerol synthetase from3 agrobacterium fabrum.
56 c3qc3B_
not modelled
82.6
14
PDB header: isomeraseChain: B: PDB Molecule: d-ribulose-5-phosphate-3-epimerase;PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
57 c5flmI_
not modelled
82.6
23
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: structure of transcribing mammalian rna polymerase ii
58 c1i3qI_
not modelled
82.3
20
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii 14.2kdPDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
59 c3hkzP_
not modelled
81.7
29
PDB header: transferaseChain: P: PDB Molecule: dna-directed rna polymerase subunit p;PDBTitle: the x-ray crystal structure of rna polymerase from archaea
60 c3cc4Z_
not modelled
81.4
28
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: co-crystal structure of anisomycin bound to the 50s ribosomal subunit
61 c3lotC_
not modelled
80.2
16
PDB header: structure genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
62 c3zf7o_
not modelled
80.2
24
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l13a, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
63 c4a17Y_
not modelled
79.9
24
PDB header: ribosomeChain: Y: PDB Molecule: rpl37a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
64 c2hnhA_
not modelled
78.7
17
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii alpha subunit;PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
65 c2qa4Z_
not modelled
78.0
28
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: a more complete structure of the the l7/l12 stalk of the2 haloarcula marismortui 50s large ribosomal subunit
66 c3floD_
not modelled
77.5
23
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast dna2 polymerase alpha in complex with its b subunit
67 c4gx9A_
not modelled
77.4
17
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii subunit epsilon,dna polymerase iiiPDBTitle: crystal structure of a dna polymerase iii alpha-epsilon chimera
68 d1ffkw_
not modelled
76.8
34
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
69 c4v36B_
not modelled
76.6
19
PDB header: transferaseChain: B: PDB Molecule: lysyl-trna-dependent l-ysyl-phosphatidylgycerol synthase;PDBTitle: the structure of l-pgs from bacillus licheniformis
70 d1muwa_
not modelled
76.5
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
71 c6k0aC_
not modelled
76.3
15
PDB header: rna binding protein/rnaChain: C: PDB Molecule: ribonuclease p protein component 3;PDBTitle: cryo-em structure of an archaeal ribonuclease p
72 c3ct7E_
not modelled
76.3
14
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
73 c4b6ap_
not modelled
75.8
21
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l17-a;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
74 c1yshD_
not modelled
75.6
24
PDB header: structural protein/rnaChain: D: PDB Molecule: ribosomal protein l37a;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
75 c3j39p_
not modelled
75.4
21
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l17;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
76 c2zkrz_
not modelled
75.3
24
PDB header: ribosomal protein/rnaChain: Z: PDB Molecule: e site t-rna;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
77 d1hjsa_
not modelled
75.2
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
78 d1jj2y_
not modelled
75.0
31
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
79 d1vqoz1
not modelled
74.7
28
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
80 c3h0gI_
not modelled
74.2
24
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
81 c3j21i_
not modelled
73.9
34
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l13p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
82 c2qkdA_
not modelled
73.7
15
PDB header: signaling protein, cell cycleChain: A: PDB Molecule: zinc finger protein zpr1;PDBTitle: crystal structure of tandem zpr1 domains
83 d7reqa2
not modelled
73.4
13
Fold: Flavodoxin-likeSuperfamily: Cobalamin (vitamin B12)-binding domainFamily: Cobalamin (vitamin B12)-binding domain
84 c3izrm_
not modelled
72.9
24
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l23 (l14p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
85 c3jyw9_
not modelled
72.6
21
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
86 c3dx5A_
not modelled
72.3
10
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
87 c5zb8B_
not modelled
71.5
14
PDB header: dna binding proteinChain: B: PDB Molecule: pfuendoq;PDBTitle: crystal structure of the novel lesion-specific endonuclease pfuendoq2 from pyrococcus furiosus
88 c5y06A_
not modelled
71.3
25
PDB header: unknown functionChain: A: PDB Molecule: msmeg_4306;PDBTitle: structural characterization of msmeg_4306 from mycobacterium smegmatis
89 c3axtA_
not modelled
70.5
17
PDB header: transferaseChain: A: PDB Molecule: probable n(2),n(2)-dimethylguanosine trna methyltransferasePDBTitle: complex structure of trna methyltransferase trm1 from aquifex aeolicus2 with s-adenosyl-l-methionine
90 c6ncsB_
not modelled
70.3
11
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-acetylneuraminic acid (sialic acid) synthetase;PDBTitle: crystal structure of n-acetylneuraminic acid (sialic acid) synthetase2 from leptospira borgpetersenii serovar hardjo-bovis in complex with3 citrate
91 c4k3zA_
not modelled
69.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
92 c2nb9A_
not modelled
69.9
32
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of zitp zinc finger
93 c5ijlA_
not modelled
69.0
27
PDB header: transferaseChain: A: PDB Molecule: dna polymerase ii large subunit;PDBTitle: d-family dna polymerase - dp2 subunit (catalytic subunit)
94 c4c2mX_
not modelled
69.0
20
PDB header: transcriptionChain: X: PDB Molecule: dna-directed rna polymerase i subunit rpa12;PDBTitle: structure of rna polymerase i at 2.8 a resolution
95 d1xima_
not modelled
68.9
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
96 d1bxca_
not modelled
68.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
97 c4l9mA_
not modelled
68.3
22
PDB header: signaling proteinChain: A: PDB Molecule: ras guanyl-releasing protein 1;PDBTitle: autoinhibited state of the ras-specific exchange factor rasgrp1
98 c4yddF_
not modelled
68.2
3
PDB header: oxidoreductaseChain: F: PDB Molecule: dmso reductase family type ii enzyme, iron-sulfur subunit;PDBTitle: crystal structure of the perchlorate reductase pcrab from azospira2 suillum ps
99 c1s1i9_
not modelled
67.8
21
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
100 c3rmjB_
not modelled
67.6
10
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
101 d1qt1a_
not modelled
67.3
17
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
102 c4ur7B_
not modelled
67.2
6
PDB header: lyaseChain: B: PDB Molecule: keto-deoxy-d-galactarate dehydratase;PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
103 c3j21e_
not modelled
67.0
35
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l5p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
104 d1yx1a1
not modelled
66.6
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like
105 c3ogrA_
not modelled
66.4
14
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
106 c3e35A_
not modelled
66.4
12
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein sco1997;PDBTitle: actinobacteria-specific protein of unknown function, sco1997
107 c5zhzA_
not modelled
66.2
15
PDB header: dna binding proteinChain: A: PDB Molecule: probable endonuclease 4;PDBTitle: crystal structure of the apurinic/apyrimidinic endonuclease iv from2 mycobacterium tuberculosis
108 c3bicA_
not modelled
65.2
12
PDB header: isomeraseChain: A: PDB Molecule: methylmalonyl-coa mutase, mitochondrial precursor;PDBTitle: crystal structure of human methylmalonyl-coa mutase
109 d1bxba_
not modelled
64.1
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
110 c1dvbA_
not modelled
63.8
35
PDB header: electron transportChain: A: PDB Molecule: rubrerythrin;PDBTitle: rubrerythrin
111 c2lcqA_
not modelled
63.4
28
PDB header: metal binding proteinChain: A: PDB Molecule: putative toxin vapc6;PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
112 c1hk8A_
not modelled
63.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity regulation in2 class iii ribonucleotide reductases: nrdd in complex with dgtp
113 d1hk8a_
not modelled
63.4
15
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit
114 c4rulA_
not modelled
63.3
29
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 1;PDBTitle: crystal structure of full-length e.coli topoisomerase i in complex2 with ssdna
115 d1ujpa_
not modelled
63.1
15
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
116 d2apob1
not modelled
63.1
28
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like
117 c2p0oA_
not modelled
63.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf871;PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
118 d2q02a1
not modelled
63.0
12
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
119 c1yuzB_
not modelled
62.7
36
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
120 c2zaeB_
not modelled
61.8
21
PDB header: hydrolaseChain: B: PDB Molecule: ribonuclease p protein component 4;PDBTitle: crystal structure of protein ph1601p in complex with protein ph1771p2 of archaeal ribonuclease p from pyrococcus horikoshii ot3