| 1 | c5lc5D_
|
|
 |
100.0 |
44 |
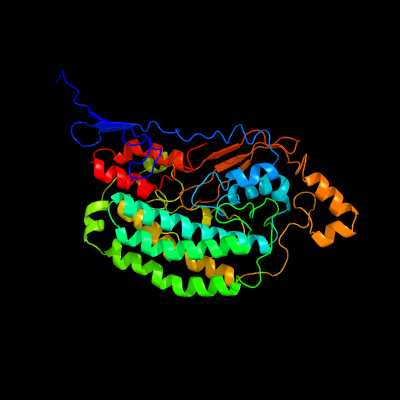
PDB header:oxidoreductase
Chain: D: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 2,
PDBTitle: structure of mammalian respiratory complex i, class2
|
|
|
|
| 2 | c6gcsC_
|
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:49-kda protein (nucm);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
|
| 3 | c6humH_
|
|
 |
100.0 |
39 |
PDB header:proton transport
Chain: H: PDB Molecule:nad(p)h-quinone oxidoreductase subunit h;
PDBTitle: structure of the photosynthetic complex i from thermosynechococcus2 elongatus
|
|
|
|
| 4 | c6cfwL_
|
|
 |
100.0 |
32 |
PDB header:membrane protein
Chain: L: PDB Molecule:membrane-bound hydrogenase subunit alpha;
PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
|
|
|
|
| 5 | d2fug41
|
|
 |
100.0 |
49 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nqo4-like |
|
|
|
| 6 | c2fug4_
|
|
 |
100.0 |
49 |
PDB header:oxidoreductase
Chain: 4: PDB Molecule:nadh-quinone oxidoreductase chain 4;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
|
|
|
| 7 | d1e3db_
|
|
 |
100.0 |
22 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nickel-iron hydrogenase, large subunit |
|
|
|
| 8 | d1frfl_
|
|
 |
100.0 |
18 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nickel-iron hydrogenase, large subunit |
|
|
|
| 9 | d1yq9h1
|
|
 |
100.0 |
21 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nickel-iron hydrogenase, large subunit |
|
|
|
| 10 | c1h2aL_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:hydrogenase;
PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
|
|
|
|
| 11 | c5aa5C_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nife-hydrogenase large subunit, hofg;
PDBTitle: actinobacterial-type nife-hydrogenase from ralstonia eutropha h16 at2 2.85 angstrom resolution
|
|
|
|
| 12 | c2wpnB_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:periplasmic [nifese] hydrogenase, large subunit,
PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase from d.2 vulgaris hildenborough
|
|
|
|
| 13 | c6ehqM_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:hydrogenase-2 large chain;
PDBTitle: e. coli hydrogenase-2 (as isolated form).
|
|
|
|
| 14 | d1cc1l_
|
|
 |
100.0 |
18 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nickel-iron hydrogenase, large subunit |
|
|
|
| 15 | c3myrB_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nickel-dependent hydrogenase large subunit;
PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
|
|
|
|
| 16 | c5odqF_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:methyl-viologen reducing hydrogenase, subunit a;
PDBTitle: heterodisulfide reductase / [nife]-hydrogenase complex from2 methanothermococcus thermolithotrophicus soaked with3 bromoethanesulfonate.
|
|
|
|
| 17 | c5xf9H_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nad-reducing hydrogenase;
PDBTitle: crystal structure of nad+-reducing [nife]-hydrogenase in the air-2 oxidized state
|
|
|
|
| 18 | d1wuil1
|
|
 |
100.0 |
19 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nickel-iron hydrogenase, large subunit |
|
|
|
| 19 | c3zfsA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:f420-reducing hydrogenase, subunit alpha;
PDBTitle: cryo-em structure of the f420-reducing nife-hydrogenase from a2 methanogenic archaeon with bound substrate
|
|
|
|
| 20 | c3useL_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:hydrogenase-1 large chain;
PDBTitle: crystal structure of e. coli hydrogenase-1 in its as-isolated form
|
|
|
|
| 21 | c4c3oC_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:hydrogenase-1 large subunit;
PDBTitle: structure and function of an oxygen tolerant nife hydrogenase from2 salmonella
|
|
|
| 22 | c5yy0A_ |
|
not modelled |
100.0 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:cytosolic nife-hydrogenase, alpha subunit;
PDBTitle: crystal structure of the hyhl-hypa complex (form ii)
|
|
|
| 23 | c3r07C_ |
|
not modelled |
63.6 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:putative lipoate-protein ligase a subunit 2;
PDBTitle: structural analysis of an archaeal lipoylation system. a bi-partite2 lipoate protein ligase and its e2 lipoyl domain from thermoplasma3 acidophilum
|
|
|
| 24 | d2g40a1 |
|
not modelled |
59.5 |
25 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:YkgG-like |
|
|
| 25 | c2g40A_ |
|
not modelled |
59.5 |
25 |
PDB header:unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of a duf162 family protein (dr_1909) from2 deinococcus radiodurans at 1.70 a resolution
|
|
|
| 26 | d1v97a4 |
|
not modelled |
46.4 |
8 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 27 | c5ymrB_ |
|
not modelled |
33.3 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:formate acetyltransferase;
PDBTitle: the crystal structure of iseg
|
|
|
| 28 | d1yeya2 |
|
not modelled |
32.4 |
4 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 29 | d2ntka1 |
|
not modelled |
32.1 |
9 |
Fold:Ntn hydrolase-like
Superfamily:Archaeal IMP cyclohydrolase PurO
Family:Archaeal IMP cyclohydrolase PurO |
|
|
| 30 | d1ffvc1 |
|
not modelled |
31.0 |
21 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 31 | d1r9da_ |
|
not modelled |
28.5 |
15 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:PFL-like |
|
|
| 32 | c4k59A_ |
|
not modelled |
26.6 |
55 |
PDB header:rna binding protein
Chain: A: PDB Molecule:rna binding protein rsmf;
PDBTitle: crystal structure of pseudomonas aeruginosa rsmf
|
|
|
| 33 | c1ufiD_ |
|
not modelled |
25.7 |
40 |
PDB header:dna binding protein
Chain: D: PDB Molecule:major centromere autoantigen b;
PDBTitle: crystal structure of the dimerization domain of human cenp-b
|
|
|
| 34 | d1ufia_ |
|
not modelled |
25.5 |
40 |
Fold:ROP-like
Superfamily:Dimerisation domain of CENP-B
Family:Dimerisation domain of CENP-B |
|
|
| 35 | c2w3zA_ |
|
not modelled |
23.6 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deacetylase;
PDBTitle: structure of a streptococcus mutans ce4 esterase
|
|
|
| 36 | d1jroa3 |
|
not modelled |
22.4 |
26 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 37 | d1vqza1 |
|
not modelled |
21.5 |
17 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:SP1160 C-terminal domain-like |
|
|
| 38 | c4lqzA_ |
|
not modelled |
19.8 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf4909 family protein (sav1798) from2 staphylococcus aureus subsp. aureus mu50 at 1.92 a resolution
|
|
|
| 39 | c5ze4A_ |
|
not modelled |
19.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase, chloroplastic;
PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
|
|
|
| 40 | c4mnoA_ |
|
not modelled |
18.0 |
15 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor 1a;
PDBTitle: crystal structure of aif1a from pyrococcus abyssi
|
|
|
| 41 | c2f3oB_ |
|
not modelled |
17.8 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:pyruvate formate-lyase 2;
PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
|
|
|
| 42 | c5j84A_ |
|
not modelled |
17.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydroxy-acid dehydratase;
PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
|
|
|
| 43 | c5dkoA_ |
|
not modelled |
16.8 |
18 |
PDB header:replication
Chain: A: PDB Molecule:cell division protein zapd;
PDBTitle: the structure of escherichia coli zapd
|
|
|
| 44 | d1rm6b1 |
|
not modelled |
15.8 |
18 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 45 | c3hrdC_ |
|
not modelled |
15.5 |
12 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinate dehydrogenase fad-subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
|
|
| 46 | c2w3rG_ |
|
not modelled |
15.5 |
26 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
|
|
| 47 | d1jdfa2 |
|
not modelled |
14.7 |
9 |
Fold:Enolase N-terminal domain-like
Superfamily:Enolase N-terminal domain-like
Family:Enolase N-terminal domain-like |
|
|
| 48 | d1u07a_ |
|
not modelled |
13.8 |
17 |
Fold:TolA/TonB C-terminal domain
Superfamily:TolA/TonB C-terminal domain
Family:TonB |
|
|
| 49 | c2dd9C_ |
|
not modelled |
13.2 |
15 |
PDB header:luminescent protein
Chain: C: PDB Molecule:green fluorescent protein;
PDBTitle: a mutant of gfp-like protein from chiridius poppei
|
|
|
| 50 | d1t3qc1 |
|
not modelled |
11.7 |
18 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 51 | d1n62c1 |
|
not modelled |
11.6 |
15 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:CO dehydrogenase flavoprotein C-terminal domain-like
Family:CO dehydrogenase flavoprotein C-terminal domain-like |
|
|
| 52 | c2dgyA_ |
|
not modelled |
11.5 |
16 |
PDB header:translation
Chain: A: PDB Molecule:mgc11102 protein;
PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
|
|
|
| 53 | d1x2ga1 |
|
not modelled |
11.3 |
17 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:SP1160 C-terminal domain-like |
|
|
| 54 | c4af3D_ |
|
not modelled |
11.0 |
13 |
PDB header:transferase/inhibitor
Chain: D: PDB Molecule:inner centromere protein;
PDBTitle: human aurora b kinase in complex with incenp and vx-680
|
|
|
| 55 | c3o6uB_ |
|
not modelled |
11.0 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein cpe2226;
PDBTitle: crystal structure of cpe2226 protein from clostridium perfringens.2 northeast structural genomics consortium target cpr195
|
|
|
| 56 | c2vgpD_ |
|
not modelled |
10.9 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:inner centromere protein a;
PDBTitle: crystal structure of aurora b kinase in complex with a2 aminothiazole inhibitor
|
|
|
| 57 | c2grxC_ |
|
not modelled |
10.9 |
17 |
PDB header:metal transport
Chain: C: PDB Molecule:protein tonb;
PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
|
|
|
| 58 | c5ibyA_ |
|
not modelled |
10.8 |
21 |
PDB header:ligase,transferase
Chain: A: PDB Molecule:lipoate--protein ligase;
PDBTitle: crystal structure of enterococcus faecalis lipoate-protein ligase a2 (lpla-2) in complex with lipoic acid
|
|
|
| 59 | c3km3B_ |
|
not modelled |
10.5 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:deoxycytidine triphosphate deaminase;
PDBTitle: crystal structure of eoxycytidine triphosphate deaminase from2 anaplasma phagocytophilum at 2.1a resolution
|
|
|
| 60 | c1wvtA_ |
|
not modelled |
10.4 |
5 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st2180;
PDBTitle: crystal structure of uncharacterized protein st2180 from sulfolobus2 tokodaii
|
|
|
| 61 | c5i2gB_ |
|
not modelled |
10.4 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:diol dehydratase;
PDBTitle: 1,2-propanediol dehydration in roseburia inulinivorans; structural2 basis for substrate and enantiomer selectivity
|
|
|
| 62 | d2p7ja1 |
|
not modelled |
10.4 |
17 |
Fold:Profilin-like
Superfamily:Sensory domain-like
Family:YkuI C-terminal domain-like |
|
|
| 63 | c6gr8B_ |
|
not modelled |
10.2 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:inner centromere protein;
PDBTitle: human aurkc incenp complex bound to brd-7880
|
|
|
| 64 | c3fa4D_ |
|
not modelled |
9.5 |
21 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 65 | c4qamB_ |
|
not modelled |
9.5 |
27 |
PDB header:signaling protein
Chain: B: PDB Molecule:x-linked retinitis pigmentosa gtpase regulator-interacting
PDBTitle: crystal structure of the rpgr rcc1-like domain in complex with the2 rpgr-interacting domain of rpgrip1
|
|
|
| 66 | c4dmbA_ |
|
not modelled |
9.2 |
13 |
PDB header:immune system
Chain: A: PDB Molecule:hd domain-containing protein 2;
PDBTitle: x-ray structure of human hepatitus c virus ns5a-transactivated protein2 2 at the resolution 1.9a, northeast structural genomics consortium3 (nesg) target hr6723
|
|
|
| 67 | c2dnwA_ |
|
not modelled |
9.0 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: solution structure of rsgi ruh-059, an acp domain of acyl2 carrier protein, mitochondrial [precursor] from human cdna
|
|
|
| 68 | c2mw4D_ |
|
not modelled |
8.9 |
24 |
PDB header:transcription
Chain: D: PDB Molecule:transcription factor protein;
PDBTitle: tetramerization domain of the ciona intestinalis p53/p73-b2 transcription factor protein
|
|
|
| 69 | d2gskb1 |
|
not modelled |
8.9 |
17 |
Fold:TolA/TonB C-terminal domain
Superfamily:TolA/TonB C-terminal domain
Family:TonB |
|
|
| 70 | c3va8A_ |
|
not modelled |
8.6 |
5 |
PDB header:lyase
Chain: A: PDB Molecule:probable dehydratase;
PDBTitle: crystal structure of enolase fg03645.1 (target efi-502278) from2 gibberella zeae ph-1 complexed with magnesium, formate and sulfate
|
|
|
| 71 | c2m1mA_ |
|
not modelled |
8.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:auxin-induced protein iaa4;
PDBTitle: solution structure of the psiaa4 oligomerization domain reveals2 interaction modes for transcription factors in early auxin response
|
|
|
| 72 | c2l9fA_ |
|
not modelled |
8.5 |
3 |
PDB header:transferase
Chain: A: PDB Molecule:cale8;
PDBTitle: nmr solution structure of meacp
|
|
|
| 73 | c2p4vA_ |
|
not modelled |
8.4 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor greb;
PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
|
|
|
| 74 | d1ik0a_ |
|
not modelled |
8.4 |
14 |
Fold:4-helical cytokines
Superfamily:4-helical cytokines
Family:Short-chain cytokines |
|
|
| 75 | c5ij6A_ |
|
not modelled |
8.1 |
11 |
PDB header:ligase,transferase
Chain: A: PDB Molecule:lipoate--protein ligase;
PDBTitle: crystal structure of enterococcus faecalis lipoate-protein ligase a2 (lpla-1) in complex with lipoic acid
|
|
|
| 76 | c2oqkA_ |
|
not modelled |
8.0 |
19 |
PDB header:translation
Chain: A: PDB Molecule:putative translation initiation factor eif-1a;
PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
|
|
|
| 77 | c5fayA_ |
|
not modelled |
7.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:choline trimethylamine-lyase;
PDBTitle: y208f mutant of choline tma-lyase
|
|
|
| 78 | c4byl5_ |
|
not modelled |
7.9 |
5 |
PDB header:ribosome
Chain: 5: PDB Molecule:ubiquitin-40s ribosomal protein s31;
PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
|
|
|
| 79 | c4uer9_ |
|
not modelled |
7.9 |
5 |
PDB header:translation
Chain: 9: PDB Molecule:es31;
PDBTitle: 40s-eif1-eif1a-eif3-eif3j translation initiation complex from2 lachancea kluyveri
|
|
|
| 80 | c3u5cf_ |
|
not modelled |
7.9 |
5 |
PDB header:ribosome
Chain: F: PDB Molecule:40s ribosomal protein s5;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome a
|
|
|
| 81 | c4byt5_ |
|
not modelled |
7.9 |
5 |
PDB header:ribosome
Chain: 5: PDB Molecule:ubiquitin-40s ribosomal protein s31;
PDBTitle: cryo-em reconstruction of the 80s-eif5b-met-itrnamet2 eukaryotic translation initiation complex
|
|
|
| 82 | c5fmtB_ |
|
not modelled |
7.8 |
17 |
PDB header:protein transport
Chain: B: PDB Molecule:flagellar associated protein;
PDBTitle: crift54 ch-domain
|
|
|
| 83 | d1o98a1 |
|
not modelled |
7.7 |
12 |
Fold:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Superfamily:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain |
|
|
| 84 | c5ikfA_ |
|
not modelled |
7.6 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:chromatin remodeling factor mit1;
PDBTitle: crystal structure of the c-terminal domain of the mit1 nucleosome2 remodeler in complex with clr1
|
|
|
| 85 | d2up1a2 |
|
not modelled |
7.6 |
21 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
|
|
| 86 | d1qxha_ |
|
not modelled |
7.5 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
|
|
| 87 | c6eacC_ |
|
not modelled |
7.5 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:selo;
PDBTitle: pseudomonas syringae selo
|
|
|
| 88 | c4tr3A_ |
|
not modelled |
7.3 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:type iii iodothyronine deiodinase;
PDBTitle: mouse iodothyronine deiodinase 3 catalytic core, semet-labeled active2 site mutant secys->cys
|
|
|
| 89 | d1rtya_ |
|
not modelled |
7.3 |
9 |
Fold:Ferritin-like
Superfamily:Cobalamin adenosyltransferase-like
Family:Cobalamin adenosyltransferase |
|
|
| 90 | c1xx3A_ |
|
not modelled |
7.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:tonb protein;
PDBTitle: solution structure of escherichia coli tonb-ctd
|
|
|
| 91 | d2oeza1 |
|
not modelled |
6.9 |
18 |
Fold:YacF-like
Superfamily:YacF-like
Family:YacF-like |
|
|
| 92 | c1rm6E_ |
|
not modelled |
6.7 |
16 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
|
|
| 93 | c4cuyf_ |
|
not modelled |
6.7 |
4 |
PDB header:ribosome
Chain: F: PDB Molecule:us7;
PDBTitle: kluyveromyces lactis 80s ribosome in complex with crpv-ires
|
|
|
| 94 | c4f4fB_ |
|
not modelled |
6.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:threonine synthase;
PDBTitle: x-ray crystal structure of plp bound threonine synthase from brucella2 melitensis
|
|
|
| 95 | c3j81i_ |
|
not modelled |
6.7 |
20 |
PDB header:ribosome
Chain: I: PDB Molecule:es8;
PDBTitle: cryoem structure of a partial yeast 48s preinitiation complex
|
|
|
| 96 | c5djnA_ |
|
not modelled |
6.7 |
30 |
PDB header:transport protein
Chain: A: PDB Molecule:kinesin-like protein;
PDBTitle: crystal structure of the kinesin-3 kif13a nc-cc1 mutant - deletion of2 p390
|
|
|
| 97 | c3sc0A_ |
|
not modelled |
6.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methylmalonic aciduria and homocystinuria type c protein;
PDBTitle: crystal structure of mmachc (1-238), a human b12 processing enzyme,2 complexed with methylcobalamin
|
|
|
| 98 | d1rtyb_ |
|
not modelled |
6.5 |
10 |
Fold:Ferritin-like
Superfamily:Cobalamin adenosyltransferase-like
Family:Cobalamin adenosyltransferase |
|
|
| 99 | c5ehbA_ |
|
not modelled |
6.5 |
35 |
PDB header:de novo protein
Chain: A: PDB Molecule:phiosyi;
PDBTitle: a de novo designed hexameric coiled-coil peptide with iodotyrosine
|
|
|