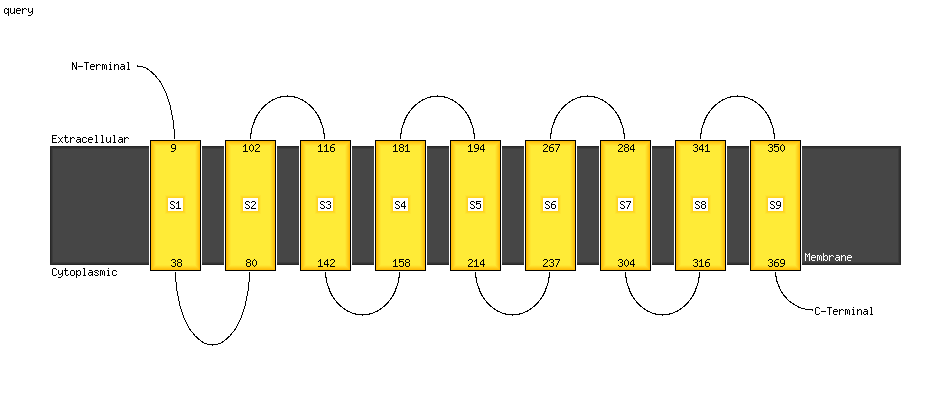
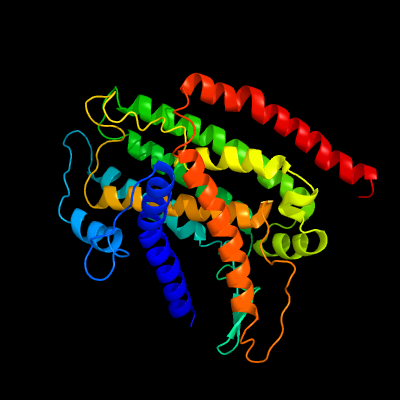
| 1 | c6gcs1_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: 1: PDB Molecule:nd1 subunit (nu1m);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
|
| 2 | c6humA_
|
|
 |
100.0 |
37 |
PDB header:proton transport
Chain: A: PDB Molecule:nad(p)h-quinone oxidoreductase subunit 1;
PDBTitle: structure of the photosynthetic complex i from thermosynechococcus2 elongatus
|
|
|
|
| 3 | c5lc5H_
|
|
 |
100.0 |
40 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nadh-ubiquinone oxidoreductase chain 1;
PDBTitle: structure of mammalian respiratory complex i, class2
|
|
|
|
| 4 | c6cfwM_
|
|
 |
100.0 |
25 |
PDB header:membrane protein
Chain: M: PDB Molecule:mbh13 nadh dehydrogenase subunit;
PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
|
|
|
|
| 5 | c4he8H_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nadh-quinone oxidoreductase subunit 8;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 thermus thermophilus
|
|
|
|
| 6 | c6g2jL_
|
|
 |
91.3 |
20 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-ubiquinone oxidoreductase chain 5;
PDBTitle: mouse mitochondrial complex i in the active state
|
|
|
|
| 7 | c6gcs5_
|
|
 |
91.1 |
15 |
PDB header:oxidoreductase
Chain: 5: PDB Molecule:nd5 subunit (nu5m);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
|
| 8 | c4heaT_
|
|
 |
90.4 |
18 |
PDB header:oxidoreductase
Chain: T: PDB Molecule:nadh-quinone oxidoreductase subunit 12;
PDBTitle: crystal structure of the entire respiratory complex i from thermus2 thermophilus
|
|
|
|
| 9 | c5ldwM_
|
|
 |
89.4 |
14 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-ubiquinone oxidoreductase chain 4;
PDBTitle: structure of mammalian respiratory complex i, class1
|
|
|
|
| 10 | c6gcs4_
|
|
 |
88.7 |
17 |
PDB header:oxidoreductase
Chain: 4: PDB Molecule:nd4 subunit (nu4m);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
|
| 11 | c6gcs2_
|
|
 |
88.3 |
7 |
PDB header:oxidoreductase
Chain: 2: PDB Molecule:nd2 subunit (nu2m);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
|
| 12 | c6cfwH_
|
|
 |
88.1 |
11 |
PDB header:membrane protein
Chain: H: PDB Molecule:monovalent cation/h+ antiporter subunit d;
PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
|
|
|
|
| 13 | c5ldwL_
|
|
 |
87.7 |
15 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-ubiquinone oxidoreductase chain 5;
PDBTitle: structure of mammalian respiratory complex i, class1
|
|
|
|
| 14 | c4he8M_
|
|
 |
87.2 |
11 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit 13;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 thermus thermophilus
|
|
|
|
| 15 | c6humF_
|
|
 |
86.4 |
15 |
PDB header:proton transport
Chain: F: PDB Molecule:nadh dehydrogenase subunit 5;
PDBTitle: structure of the photosynthetic complex i from thermosynechococcus2 elongatus
|
|
|
|
| 16 | c3rkoL_
|
|
 |
79.0 |
13 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
|
|
|
|
| 17 | c6humB_
|
|
 |
72.0 |
12 |
PDB header:proton transport
Chain: B: PDB Molecule:nad(p)h-quinone oxidoreductase subunit 2;
PDBTitle: structure of the photosynthetic complex i from thermosynechococcus2 elongatus
|
|
|
|
| 18 | c5xtdi_
|
|
 |
50.2 |
9 |
PDB header:oxidoreductase/electron transport
Chain: I: PDB Molecule:nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunit
PDBTitle: cryo-em structure of human respiratory complex i
|
|
|
|
| 19 | c2jpkA_
|
|
 |
33.7 |
23 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in dpc
|
|
|
|
| 20 | c4p6vE_
|
|
 |
33.4 |
12 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:na(+)-translocating nadh-quinone reductase subunit e;
PDBTitle: crystal structure of the na+-translocating nadh: ubiquinone2 oxidoreductase from vibrio cholerae
|
|
|
|
| 21 | d2a65a1 |
|
not modelled |
25.3 |
10 |
Fold:SNF-like
Superfamily:SNF-like
Family:SNF-like |
|
|
| 22 | c2jpmA_ |
|
not modelled |
21.1 |
23 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in tfe
|
|
|
| 23 | c5ldwN_ |
|
not modelled |
20.0 |
8 |
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-ubiquinone oxidoreductase chain 2;
PDBTitle: structure of mammalian respiratory complex i, class1
|
|
|
| 24 | c2kncA_ |
|
not modelled |
19.7 |
22 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
|
|
| 25 | d1xn8a_ |
|
not modelled |
18.8 |
19 |
Fold:Hypothetical protein YqbG
Superfamily:Hypothetical protein YqbG
Family:Hypothetical protein YqbG |
|
|
| 26 | d1jb0l_ |
|
not modelled |
16.8 |
20 |
Fold:Photosystem I reaction center subunit XI, PsaL
Superfamily:Photosystem I reaction center subunit XI, PsaL
Family:Photosystem I reaction center subunit XI, PsaL |
|
|
| 27 | c1wa7B_ |
|
not modelled |
14.5 |
60 |
PDB header:sh3 domain
Chain: B: PDB Molecule:hypothetical 28.7 kda protein in dhfr 3'region
PDBTitle: sh3 domain of human lyn tyrosine kinase in complex with a2 herpesviral ligand
|
|
|
| 28 | c4djiA_ |
|
not modelled |
11.9 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:probable glutamate/gamma-aminobutyrate antiporter;
PDBTitle: structure of glutamate-gaba antiporter gadc
|
|
|
| 29 | c2k1aA_ |
|
not modelled |
10.2 |
22 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
|
|
|
| 30 | c3pesA_ |
|
not modelled |
9.7 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein gp49;
PDBTitle: crystal structure of uncharacterized protein from pseudomonas phage2 yua
|
|
|
| 31 | c5cwwC_ |
|
not modelled |
9.0 |
61 |
PDB header:transport protein
Chain: C: PDB Molecule:nucleoporin nup159;
PDBTitle: crystal structure of the chaetomium thermophilum heterotrimeric nup822 ntd-nup159 tail-nup145n apd complex
|
|
|
| 32 | c2vxsB_ |
|
not modelled |
8.5 |
36 |
PDB header:cytokine
Chain: B: PDB Molecule:interleukin-17a;
PDBTitle: structure of il-17a in complex with a potent, fully human2 neutralising antibody
|
|
|
| 33 | c4q2uM_ |
|
not modelled |
8.2 |
24 |
PDB header:toxin/toxin repressor
Chain: M: PDB Molecule:antitoxin dinj;
PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
|
|
|
| 34 | d1ufra_ |
|
not modelled |
7.8 |
20 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
|
|
| 35 | c4he8I_ |
|
not modelled |
7.5 |
12 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:nadh-quinone oxidoreductase subunit 14;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 thermus thermophilus
|
|
|
| 36 | c5aexJ_ |
|
not modelled |
7.2 |
17 |
PDB header:membrane protein
Chain: J: PDB Molecule:ammonium transporter mep2;
PDBTitle: crystal structure of saccharomyces cerevisiae mep2
|
|
|
| 37 | c6ahzA_ |
|
not modelled |
7.1 |
28 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:cmp-n-acetylneuraminate-poly-alpha-2,8-sialyltransferase;
PDBTitle: the nmr structure of the polysialyltranseferase domain (pstd) in2 polysialyltransferase st8siaiv
|
|
|
| 38 | c2l8sA_ |
|
not modelled |
6.9 |
30 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-1;
PDBTitle: solution nmr structure of transmembrane and cytosolic regions of2 integrin alpha1 in detergent micelles
|
|
|
| 39 | d1e6va1 |
|
not modelled |
6.8 |
30 |
Fold:Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Superfamily:Methyl-coenzyme M reductase alpha and beta chain C-terminal domain
Family:Methyl-coenzyme M reductase alpha and beta chain C-terminal domain |
|
|
| 40 | c4h25C_ |
|
not modelled |
6.8 |
57 |
PDB header:immune system
Chain: C: PDB Molecule:peptide;
PDBTitle: tcr interaction with peptide mimics of nickel offers structure2 insights to nickel contact allergy
|
|
|
| 41 | c4h25F_ |
|
not modelled |
6.8 |
57 |
PDB header:immune system
Chain: F: PDB Molecule:peptide;
PDBTitle: tcr interaction with peptide mimics of nickel offers structure2 insights to nickel contact allergy
|
|
|
| 42 | c3llbA_ |
|
not modelled |
6.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
|
|
|
| 43 | d2f05a1 |
|
not modelled |
6.4 |
21 |
Fold:PAH2 domain
Superfamily:PAH2 domain
Family:PAH2 domain |
|
|
| 44 | c2a2bA_ |
|
not modelled |
6.4 |
60 |
PDB header:antibiotic
Chain: A: PDB Molecule:bacteriocin curvacin a;
PDBTitle: curvacin a
|
|
|
| 45 | d1tcaa_ |
|
not modelled |
6.4 |
15 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
|
|
| 46 | d1un8a4 |
|
not modelled |
6.3 |
26 |
Fold:DAK1/DegV-like
Superfamily:DAK1/DegV-like
Family:DAK1 |
|
|
| 47 | d1s5qb_ |
|
not modelled |
6.3 |
22 |
Fold:PAH2 domain
Superfamily:PAH2 domain
Family:PAH2 domain |
|
|
| 48 | c3qilM_ |
|
not modelled |
6.2 |
26 |
PDB header:structural protein
Chain: M: PDB Molecule:clathrin heavy chain 1;
PDBTitle: crystal structure analysis of the clathrin trimerization domain
|
|
|
| 49 | d1jpya_ |
|
not modelled |
6.1 |
36 |
Fold:Cystine-knot cytokines
Superfamily:Cystine-knot cytokines
Family:Interleukin 17F, IL-17F |
|
|
| 50 | c3etoB_ |
|
not modelled |
6.1 |
15 |
PDB header:signaling protein
Chain: B: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: 2 angstrom xray structure of the notch1 negative regulatory region2 (nrr)
|
|
|
| 51 | c3m3nW_ |
|
not modelled |
6.0 |
41 |
PDB header:structural protein
Chain: W: PDB Molecule:neural wiskott-aldrich syndrome protein;
PDBTitle: structure of a longitudinal actin dimer assembled by tandem w domains
|
|
|
| 52 | c1q69B_ |
|
not modelled |
5.9 |
36 |
PDB header:membrane protein/transferase
Chain: B: PDB Molecule:proto-oncogene tyrosine-protein kinase lck;
PDBTitle: solution structure of t-cell surface glycoprotein cd8 alpha2 chain and proto-oncogene tyrosine-protein kinase lck3 fragments
|
|
|
| 53 | c1q68B_ |
|
not modelled |
5.9 |
36 |
PDB header:membrane protein/transferase
Chain: B: PDB Molecule:proto-oncogene tyrosine-protein kinase lck;
PDBTitle: solution structure of t-cell surface glycoprotein cd4 and2 proto-oncogene tyrosine-protein kinase lck fragments
|
|
|
| 54 | c2n07X_ |
|
not modelled |
5.7 |
67 |
PDB header:toxin
Chain: X: PDB Molecule:alpha-conotoxin vc1a;
PDBTitle: design of a highly stable disulfide-deleted mutant of analgesic cyclic2 alpha-conotoxin vc1.1
|
|
|
| 55 | c3dyuB_ |
|
not modelled |
5.7 |
40 |
PDB header:transport protein
Chain: B: PDB Molecule:sorting nexin-9;
PDBTitle: crystal structure of snx9px-bar (230-595), h32
|
|
|
| 56 | c2dbhA_ |
|
not modelled |
5.7 |
36 |
PDB header:signaling protein
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily
PDBTitle: solution structure of the carboxyl-terminal card-like2 domain in human tnfr-related death receptor-6
|
|
|
| 57 | d2r2za1 |
|
not modelled |
5.6 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
|
|
| 58 | c2ncyA_ |
|
not modelled |
5.6 |
33 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:pseudin-2;
PDBTitle: solution structure of pseudin-2 analog (ps-p)
|
|
|
| 59 | c2jz8A_ |
|
not modelled |
5.5 |
31 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bh09830;
PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
|
|
|
| 60 | c4qhuC_ |
|
not modelled |
5.4 |
36 |
PDB header:immune system
Chain: C: PDB Molecule:interleukin-17a;
PDBTitle: crystal structure of il-17a/fab6785 complex
|
|
|
| 61 | c1q2iA_ |
|
not modelled |
5.3 |
40 |
PDB header:antitumor protein
Chain: A: PDB Molecule:pnc27;
PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
|
|
|
| 62 | c3lygA_ |
|
not modelled |
5.1 |
50 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ntf2-like protein of unknown function;
PDBTitle: crystal structure of ntf2-like protein of unknown function2 (yp_270605.1) from colwellia psychrerythraea 34h at 1.61 a resolution
|
|
|
| 63 | d1iyka2 |
|
not modelled |
5.1 |
25 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-myristoyl transferase, NMT |
|
|
| 64 | c2rajA_ |
|
not modelled |
5.0 |
40 |
PDB header:structural protein
Chain: A: PDB Molecule:sorting nexin-9;
PDBTitle: so4 bound px-bar membrane remodeling unit of sorting nexin 9
|
|
|
| 65 | d2e9xa1 |
|
not modelled |
5.0 |
24 |
Fold:GINS helical bundle-like
Superfamily:GINS helical bundle-like
Family:PSF1 N-terminal domain-like |
|
|