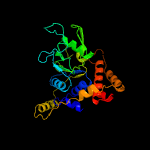
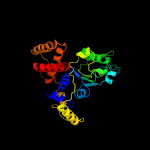
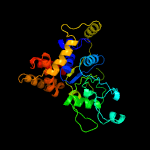
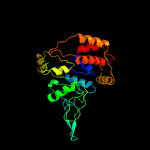
1 c2r44A_
100.0
42
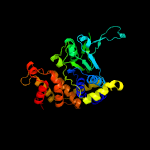
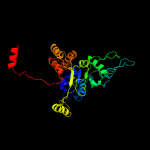
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative atpase (chu_0153) from cytophaga2 hutchinsonii atcc 33406 at 2.00 a resolution
2 c3nbxX_
100.0
21
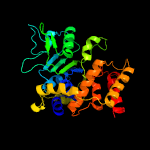
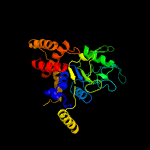
PDB header: hydrolaseChain: X: PDB Molecule: atpase rava;PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
3 c4r7zB_
100.0
20
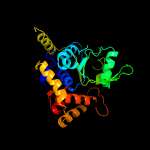
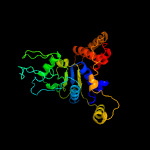
PDB header: hydrolaseChain: B: PDB Molecule: cell division control protein 21;PDBTitle: pfmcm-aaa double-octamer
4 c3ja87_
100.0
21
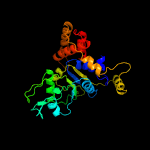
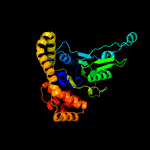
PDB header: hydrolaseChain: 7: PDB Molecule: minichromosome maintenance 7;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
5 c5udb7_
100.0
20
PDB header: replicationChain: 7: PDB Molecule: dna replication licensing factor mcm7;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
6 c3ja82_
100.0
17
PDB header: hydrolaseChain: 2: PDB Molecule: minichromosome maintenance 2;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
7 c5h7i7_
100.0
20
PDB header: hydrolaseChain: 7: PDB Molecule: dna replication licensing factor mcm7;PDBTitle: cryo-em structure of the cdt1-mcm2-7 complex in amppnp state
8 d1g8pa_
100.0
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
9 c5udb3_
100.0
17
PDB header: replicationChain: 3: PDB Molecule: dna replication licensing factor mcm3;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
10 c3ja84_
100.0
19
PDB header: hydrolaseChain: 4: PDB Molecule: minichromosome maintenance 4;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
11 c3f8tA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: predicted atpase involved in replication control, cdc46/mcmPDBTitle: crystal structure analysis of a full-length mcm homolog from2 methanopyrus kandleri
12 c5u8s4_
100.0
18
PDB header: replicationChain: 4: PDB Molecule: dna replication licensing factor mcm4;PDBTitle: structure of eukaryotic cmg helicase at a replication fork
13 c3jc72_
100.0
17
PDB header: hydrolaseChain: 2: PDB Molecule: dna replication licensing factor mcm2;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
14 c3ja83_
100.0
17
PDB header: hydrolaseChain: 3: PDB Molecule: minichromosome maintenance 3;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
15 c5v8f3_
100.0
17
PDB header: replicationChain: 3: PDB Molecule: dna replication licensing factor mcm3;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
16 c3jc76_
100.0
17
PDB header: hydrolaseChain: 6: PDB Molecule: dna replication licensing factor mcm6;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
17 c3ja85_
100.0
16
PDB header: hydrolaseChain: 5: PDB Molecule: minichromosome maintenance 5;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
18 c3ja86_
100.0
17
PDB header: hydrolaseChain: 6: PDB Molecule: minichromosome maintenance 6;PDBTitle: cryo-em structure of the mcm2-7 double hexamer
19 c6hv92_
100.0
17
PDB header: dna binding proteinChain: 2: PDB Molecule: dna replication licensing factor mcm2;PDBTitle: s. cerevisiae cmg-pol epsilon-dna
20 c5udb5_
100.0
16
PDB header: replicationChain: 5: PDB Molecule: minichromosome maintenance protein 5;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
21 c3jc57_
not modelled
100.0
21
PDB header: hydrolaseChain: 7: PDB Molecule: dna replication licensing factor mcm7;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
22 c6hv93_
not modelled
100.0
17
PDB header: dna binding proteinChain: 3: PDB Molecule: dna replication licensing factor mcm3;PDBTitle: s. cerevisiae cmg-pol epsilon-dna
23 c6hv96_
not modelled
100.0
17
PDB header: dna binding proteinChain: 6: PDB Molecule: dna replication licensing factor mcm6;PDBTitle: s. cerevisiae cmg-pol epsilon-dna
24 c3f9vA_
not modelled
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: minichromosome maintenance protein mcm;PDBTitle: crystal structure of a near full-length archaeal mcm: functional2 insights for an aaa+ hexameric helicase
25 c3jc55_
not modelled
100.0
16
PDB header: hydrolaseChain: 5: PDB Molecule: minichromosome maintenance protein 5;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
26 c3jc73_
not modelled
100.0
17
PDB header: hydrolaseChain: 3: PDB Molecule: dna replication licensing factor mcm3;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
27 c6hv97_
not modelled
100.0
22
PDB header: dna binding proteinChain: 7: PDB Molecule: dna replication licensing factor mcm7;PDBTitle: s. cerevisiae cmg-pol epsilon-dna
28 c3jc54_
not modelled
99.9
21
PDB header: hydrolaseChain: 4: PDB Molecule: dna replication licensing factor mcm4;PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
29 c3k1jA_
not modelled
99.9
23
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent protease lon;PDBTitle: crystal structure of lon protease from thermococcus onnurineus na1
30 c2c9oA_
not modelled
99.9
22
PDB header: hydrolaseChain: A: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
31 c4ww4A_
not modelled
99.9
25
PDB header: hydrolaseChain: A: PDB Molecule: ruvb-like 1;PDBTitle: double-heterohexameric rings of full-length rvb1(adp)/rvb2(adp)
32 c6i26A_
not modelled
99.9
24
PDB header: motor proteinChain: A: PDB Molecule: midasin,midasin,midasin,midasin;PDBTitle: rea1 wild type amppnp state
33 d1g41a_
not modelled
99.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
34 d1r6bx3
not modelled
99.9
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
35 c6nyyC_
not modelled
99.9
18
PDB header: translocaseChain: C: PDB Molecule: afg3-like protein 2;PDBTitle: human m-aaa protease afg3l2, substrate-bound
36 d1um8a_
not modelled
99.9
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
37 c5g4gF_
not modelled
99.9
21
PDB header: hydrolaseChain: F: PDB Molecule: vcp-like atpase;PDBTitle: structure of the atpgs-bound vat complex
38 c1r6bX_
not modelled
99.9
18
PDB header: hydrolaseChain: X: PDB Molecule: clpa protein;PDBTitle: high resolution crystal structure of clpa
39 d1ofha_
not modelled
99.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
40 c6em8F_
not modelled
99.9
22
PDB header: chaperoneChain: F: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
41 c6azyA_
not modelled
99.9
15
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp104;PDBTitle: crystal structure of hsp104 r328m/r757m mutant from calcarisporiella2 thermophila
42 c3uk6H_
not modelled
99.9
20
PDB header: hydrolaseChain: H: PDB Molecule: ruvb-like 2;PDBTitle: crystal structure of the tip48 (tip49b) hexamer
43 c5kneF_
not modelled
99.9
20
PDB header: chaperoneChain: F: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
44 c3pfiB_
not modelled
99.9
22
PDB header: hydrolaseChain: B: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 2.7 angstrom resolution crystal structure of a probable holliday2 junction dna helicase (ruvb) from campylobacter jejuni subsp. jejuni3 nctc 11168 in complex with adenosine-5'-diphosphate
45 c6blbA_
not modelled
99.9
24
PDB header: hydrolaseChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 1.88 angstrom resolution crystal structure holliday junction atp-2 dependent dna helicase (ruvb) from pseudomonas aeruginosa in complex3 with adp
46 c1qvrB_
not modelled
99.9
18
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
47 c5e7pA_
not modelled
99.9
22
PDB header: hydrolaseChain: A: PDB Molecule: cell division control protein cdc48;PDBTitle: crystal structure of msmeg_0858 (uniprot a0qqs4), a aaa atpase.
48 c5kneD_
not modelled
99.9
16
PDB header: chaperoneChain: D: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
49 d1qvra3
not modelled
99.9
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
50 c5oafB_
not modelled
99.9
26
PDB header: gene regulationChain: B: PDB Molecule: ruvb-like 2;PDBTitle: human rvb1/rvb2 heterohexamer in ino80 complex
51 c6nyyA_
not modelled
99.9
18
PDB header: translocaseChain: A: PDB Molecule: afg3-like protein 2;PDBTitle: human m-aaa protease afg3l2, substrate-bound
52 c5vq9D_
not modelled
99.9
17
PDB header: protein bindingChain: D: PDB Molecule: pachytene checkpoint protein 2 homolog;PDBTitle: structure of human trip13, apo form
53 c2dhrC_
not modelled
99.9
23
PDB header: hydrolaseChain: C: PDB Molecule: ftsh;PDBTitle: whole cytosolic region of atp-dependent metalloprotease2 ftsh (g399l)
54 c6epcJ_
not modelled
99.9
22
PDB header: hydrolaseChain: J: PDB Molecule: 26s proteasome regulatory subunit 8;PDBTitle: ground state 26s proteasome (gs2)
55 c6epdM_
not modelled
99.9
22
PDB header: hydrolaseChain: M: PDB Molecule: 26s proteasome regulatory subunit 6a;PDBTitle: substrate processing state 26s proteasome (sps1)
56 c3hteC_
not modelled
99.9
16
PDB header: motor proteinChain: C: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: crystal structure of nucleotide-free hexameric clpx
57 c4b4tH_
not modelled
99.9
16
PDB header: hydrolaseChain: H: PDB Molecule: 26s protease regulatory subunit 7 homolog;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
58 c4ciuA_
not modelled
99.9
17
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein clpb;PDBTitle: crystal structure of e. coli clpb
59 c3cf1C_
not modelled
99.9
16
PDB header: transport proteinChain: C: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: structure of p97/vcp in complex with adp/adp.alfx
60 c4b4tI_
not modelled
99.9
21
PDB header: hydrolaseChain: I: PDB Molecule: 26s protease regulatory subunit 4 homolog;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
61 c6qi8E_
not modelled
99.9
22
PDB header: chaperoneChain: E: PDB Molecule: ruvb-like 2;PDBTitle: truncated human r2tp complex, structure 3 (adp-filled)
62 c2c9oC_
not modelled
99.9
19
PDB header: hydrolaseChain: C: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
63 c6e111_
not modelled
99.9
18
PDB header: protein transportChain: 1: PDB Molecule: heat shock protein 101;PDBTitle: ptex core complex in the resetting (compact) state
64 c4b4tL_
not modelled
99.9
16
PDB header: hydrolaseChain: L: PDB Molecule: 26s protease subunit rpt4;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
65 c4b4tJ_
not modelled
99.9
19
PDB header: hydrolaseChain: J: PDB Molecule: 26s protease regulatory subunit 8 homolog;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
66 c6genX_
not modelled
99.8
24
PDB header: nuclear proteinChain: X: PDB Molecule: ruvb-like protein 1;PDBTitle: chromatin remodeller-nucleosome complex at 4.5 a resolution.
67 c5c3cB_
not modelled
99.8
19
PDB header: protein bindingChain: B: PDB Molecule: cbbq/nirq/norq domain protein;PDBTitle: structural characterization of a newly identified component of alpha-2 carboxysomes: the aaa+ domain protein cso-cbbq
68 c4d2qC_
not modelled
99.8
18
PDB header: chaperoneChain: C: PDB Molecule: clpb;PDBTitle: negative-stain electron microscopy of e. coli clpb mutant e432a (bap2 form bound to clpp)
69 c5vy9C_
not modelled
99.8
22
PDB header: chaperoneChain: C: PDB Molecule: heat shock protein 104;PDBTitle: s. cerevisiae hsp104:casein complex, middle domain conformation
70 c6em8E_
not modelled
99.8
22
PDB header: chaperoneChain: E: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
71 c6edoA_
not modelled
99.8
22
PDB header: motor proteinChain: A: PDB Molecule: midasin;PDBTitle: full-length s. pombe mdn1 in the presence of amppnp (ring region)
72 c6em8H_
not modelled
99.8
22
PDB header: chaperoneChain: H: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
73 c6orbA_
not modelled
99.8
23
PDB header: motor proteinChain: A: PDB Molecule: midasin;PDBTitle: full-length s. pombe mdn1 in the presence of atp and rbin-1
74 c4b4tK_
not modelled
99.8
22
PDB header: hydrolaseChain: K: PDB Molecule: 26s protease regulatory subunit 6b homolog;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
75 c4b4tM_
not modelled
99.8
21
PDB header: hydrolaseChain: M: PDB Molecule: 26s protease regulatory subunit 6a;PDBTitle: near-atomic resolution structural model of the yeast 26s proteasome
76 c1ojlD_
not modelled
99.8
18
PDB header: response regulatorChain: D: PDB Molecule: transcriptional regulatory protein zrar;PDBTitle: crystal structure of a sigma54-activator suggests the mechanism for2 the conformational switch necessary for sigma54 binding
77 c6djuA_
not modelled
99.8
18
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein clpb;PDBTitle: mtb clpb in complex with atpgammas and casein, conformer 1
78 c5d4wB_
not modelled
99.8
17
PDB header: chaperoneChain: B: PDB Molecule: putative heat shock protein;PDBTitle: crystal structure of hsp104
79 c5kzfJ_
not modelled
99.8
18
PDB header: hydrolaseChain: J: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of near full-length hexameric mycobacterium2 tuberculosis proteasomal atpase mpa in apo form
80 c6matE_
not modelled
99.8
18
PDB header: ribosomal proteinChain: E: PDB Molecule: rix7 mutant;PDBTitle: cryo-em structure of the essential ribosome assembly aaa-atpase rix7
81 c6hecH_
not modelled
99.8
22
PDB header: hydrolaseChain: H: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: pan-proteasome in state 4
82 c5mpaL_
not modelled
99.8
18
PDB header: hydrolaseChain: L: PDB Molecule: 26s protease subunit rpt4;PDBTitle: 26s proteasome in presence of atp (s2)
83 c1in8A_
not modelled
99.8
24
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction dna helicase ruvb;PDBTitle: thermotoga maritima ruvb t158v
84 c2ce7B_
not modelled
99.8
20
PDB header: cell division proteinChain: B: PDB Molecule: cell division protein ftsh;PDBTitle: edta treated
85 c5gjqL_
not modelled
99.8
17
PDB header: hydrolaseChain: L: PDB Molecule: 26s protease regulatory subunit 10b;PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
86 c4ypnA_
not modelled
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: lon protease;PDBTitle: crystal structure of a lona fragment containing the 3-helix bundle and2 the aaa-alpha/beta domain
87 c5t0gA_
not modelled
99.8
19
PDB header: hydrolaseChain: A: PDB Molecule: 26s protease regulatory subunit 7;PDBTitle: structural basis for dynamic regulation of the human 26s proteasome
88 c5ifwB_
not modelled
99.8
15
PDB header: signaling proteinChain: B: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: quantitative interaction mapping reveals an extended ubiquitin2 regulatory domain in aspl that disrupts functional p97 hexamers and3 induces cell death
89 c6az0A_
not modelled
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: mitochondrial inner membrane i-aaa protease supercomplexPDBTitle: mitochondrial atpase protease yme1
90 d1ixsb2
not modelled
99.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
91 c5ep4A_
not modelled
99.8
11
PDB header: transcriptionChain: A: PDB Molecule: putative repressor protein luxo;PDBTitle: structure, regulation, and inhibition of the quorum-sensing signal2 integrator luxo
92 c4ww4B_
not modelled
99.8
24
PDB header: hydrolaseChain: B: PDB Molecule: ruvb-like 2;PDBTitle: double-heterohexameric rings of full-length rvb1(adp)/rvb2(adp)
93 d1in4a2
not modelled
99.8
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
94 c4z8xC_
not modelled
99.8
22
PDB header: hydrolaseChain: C: PDB Molecule: atp-dependent zinc metalloprotease ftsh;PDBTitle: truncated ftsh from a. aeolicus
95 c3pxiB_
not modelled
99.8
18
PDB header: protein bindingChain: B: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca108:clpc
96 c3hu2C_
not modelled
99.8
26
PDB header: transport proteinChain: C: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: structure of p97 n-d1 r86a mutant in complex with atpgs
97 c3b9pA_
not modelled
99.8
23
PDB header: hydrolaseChain: A: PDB Molecule: cg5977-pa, isoform a;PDBTitle: spastin
98 c3cf2B_
not modelled
99.8
17
PDB header: transport proteinChain: B: PDB Molecule: transitional endoplasmic reticulum atpase;PDBTitle: structure of p97/vcp in complex with adp/amp-pnp
99 c4lcbA_
not modelled
99.8
21
PDB header: protein transportChain: A: PDB Molecule: cell division protein cdvc, vps4;PDBTitle: structure of vps4 homolog from acidianus hospitalis
100 c3h4mC_
not modelled
99.8
23
PDB header: hydrolaseChain: C: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: aaa atpase domain of the proteasome- activating nucleotidase
101 c4l16A_
not modelled
99.8
25
PDB header: hydrolaseChain: A: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of figl-1 aaa domain in complex with adp
102 c5ubvB_
not modelled
99.8
16
PDB header: hydrolaseChain: B: PDB Molecule: atpase domain of i-aaa protease;PDBTitle: atpase domain of i-aaa protease from myceliophthora thermophila
103 d2ce7a2
not modelled
99.8
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
104 c3vfdA_
not modelled
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: spastin;PDBTitle: human spastin aaa domain
105 c4xguB_
not modelled
99.8
20
PDB header: atp-binding proteinChain: B: PDB Molecule: putative pachytene checkpoint protein 2;PDBTitle: structure of c. elegans pch-2
106 c3eihB_
not modelled
99.8
18
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of s.cerevisiae vps4 in the presence of atpgammas
107 c6hz4B_
not modelled
99.8
18
PDB header: dna binding proteinChain: B: PDB Molecule: 5-methylcytosine-specific restriction enzyme b;PDBTitle: structure of mcrbc without dna binding domains (one half of the full2 complex)
108 c1xwiA_
not modelled
99.8
19
PDB header: protein transportChain: A: PDB Molecule: skd1 protein;PDBTitle: crystal structure of vps4b
109 d1ny5a2
not modelled
99.8
18
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
110 c6i27A_
not modelled
99.8
20
PDB header: motor proteinChain: A: PDB Molecule: midasin,midasin,midasin,midasin,midasin,midasin,midasin;PDBTitle: rea1 aaa2l-h2alpha deletion mutant in amppnp state
111 c3d8bB_
not modelled
99.8
18
PDB header: hydrolaseChain: B: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of human fidgetin-like protein 1 in complex with adp
112 d1r6bx2
not modelled
99.8
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
113 c5exsA_
not modelled
99.8
19
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator fleq;PDBTitle: aaa+ atpase fleq from pseudomonas aeruginosa bound to atp-gamma-s
114 c6b5cA_
not modelled
99.8
19
PDB header: cell cycleChain: A: PDB Molecule: katanin p60 atpase-containing subunit a-like 1;PDBTitle: structural basis for katanin self-assembly
115 c3pvsA_
not modelled
99.8
24
PDB header: recombinationChain: A: PDB Molecule: replication-associated recombination protein a;PDBTitle: structure and biochemical activities of escherichia coli mgsa
116 c5kneA_
not modelled
99.8
23
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
117 c1ojlF_
not modelled
99.8
19
PDB header: response regulatorChain: F: PDB Molecule: transcriptional regulatory protein zrar;PDBTitle: crystal structure of a sigma54-activator suggests the mechanism for2 the conformational switch necessary for sigma54 binding
118 c1hqcB_
not modelled
99.8
22
PDB header: hydrolaseChain: B: PDB Molecule: ruvb;PDBTitle: structure of ruvb from thermus thermophilus hb8
119 c2c99A_
not modelled
99.8
17
PDB header: transcription regulationChain: A: PDB Molecule: psp operon transcriptional activator;PDBTitle: structural basis of the nucleotide driven conformational2 changes in the aaa domain of transcription activator pspf
120 c2x8aA_
not modelled
99.8
20
PDB header: nuclear proteinChain: A: PDB Molecule: nuclear valosin-containing protein-like;PDBTitle: human nuclear valosin containing protein like (nvl), c-2 terminal aaa-atpase domain