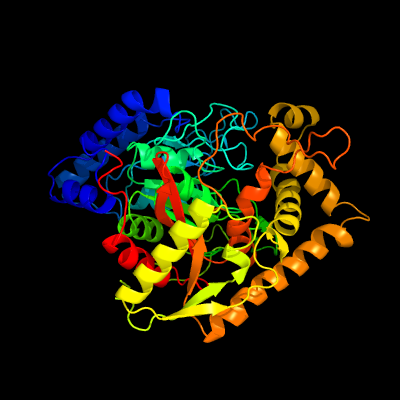
| 1 | d2f2aa1
|
|
 |
100.0 |
25 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
|
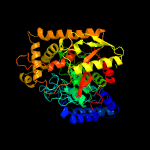
| 2 | c6diiH_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: H: PDB Molecule:fatty acid amide hydrolase;
PDBTitle: structure of arabidopsis fatty acid amide hydrolase in complex with2 methyl linolenyl fluorophosphonate
|
|
|
|
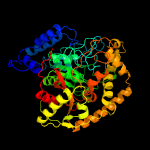
| 3 | c4wj3A_
|
|
 |
100.0 |
30 |
PDB header:ligase/rna
Chain: A: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit a;
PDBTitle: crystal structure of the asparagine transamidosome from pseudomonas2 aeruginosa
|
|
|
|
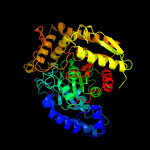
| 4 | c3h0rP_
|
|
 |
100.0 |
27 |
PDB header:ligase
Chain: P: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit a;
PDBTitle: structure of trna-dependent amidotransferase gatcab from2 aquifex aeolicus
|
|
|
|
| 5 | c4yj6A_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:aryl acylamidase;
PDBTitle: the crystal structure of a bacterial aryl acylamidase belonging to the2 amidase signature (as) enzymes family
|
|
|
|
| 6 | d1m22a_
|
|
 |
100.0 |
28 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
|
| 7 | c3a2qA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-aminohexanoate-cyclic-dimer hydrolase;
PDBTitle: structure of 6-aminohexanoate cyclic dimer hydrolase complexed with2 substrate
|
|
|
|
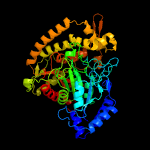
| 8 | c4gysA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:allophanate hydrolase;
PDBTitle: granulibacter bethesdensis allophanate hydrolase co-crystallized with2 malonate
|
|
|
|
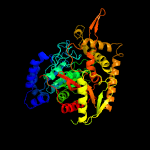
| 9 | c3kfuE_
|
|
 |
100.0 |
31 |
PDB header:ligase/rna
Chain: E: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit a;
PDBTitle: crystal structure of the transamidosome
|
|
|
|
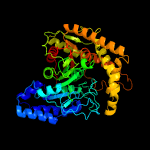
| 10 | c5h6sB_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidase;
PDBTitle: crystal structure of hydrazidase s179a mutant complexed with a2 substrate
|
|
|
|
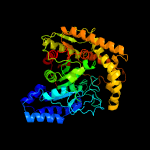
| 11 | c5h6tB_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:amidase;
PDBTitle: crystal structure of hydrazidase from microbacterium sp. strain hm58-2
|
|
|
|
| 12 | d1mt5a_
|
|
 |
100.0 |
23 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
|
| 13 | c4cp8C_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:allophanate hydrolase;
PDBTitle: structure of the amidase domain of allophanate hydrolase2 from pseudomonas sp strain adp
|
|
|
|
| 14 | c2vyaB_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:fatty-acid amide hydrolase 1;
PDBTitle: crystal structure of fatty acid amide hydrolase conjugated2 with the drug-like inhibitor pf-750
|
|
|
|
| 15 | c6c6gA_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:biuret hydrolase;
PDBTitle: an unexpected vestigial protein complex reveals the evolutionary2 origins of an s-triazine catabolic enzyme. inhibitor bound complex.
|
|
|
|
| 16 | c2dc0A_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable amidase;
PDBTitle: crystal structure of amidase
|
|
|
|
| 17 | d2gi3a1
|
|
 |
100.0 |
25 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
|
| 18 | c3a1iA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidase;
PDBTitle: crystal structure of rhodococcus sp. n-771 amidase complexed2 with benzamide
|
|
|
|
| 19 | d1ocka_
|
|
 |
100.0 |
26 |
Fold:Amidase signature (AS) enzymes
Superfamily:Amidase signature (AS) enzymes
Family:Amidase signature (AS) enzymes |
|
|
|
| 20 | c4issA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:allophanate hydrolase;
PDBTitle: semet-substituted kluyveromyces lactis allophanate hydrolase
|
|
|
|
| 21 | c5i8iD_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:urea amidolyase;
PDBTitle: crystal structure of the k. lactis urea amidolyase
|
|
|
| 22 | c5ewqC_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:amidase;
PDBTitle: the crystal structure of an amidase family protein from bacillus2 anthracis str. ames
|
|
|
| 23 | c4n0hA_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit a,
PDBTitle: crystal structure of s. cerevisiae mitochondrial gatfab
|
|
|
| 24 | c3r4kD_ |
|
not modelled |
56.5 |
20 |
PDB header:dna binding protein
Chain: D: PDB Molecule:transcriptional regulator, iclr family;
PDBTitle: crystal structure of a putative iclr transcriptional regulator2 (tm1040_3717) from silicibacter sp. tm1040 at 2.46 a resolution
|
|
|
| 25 | d1h5wa_ |
|
not modelled |
50.7 |
15 |
Fold:Upper collar protein gp10 (connector protein)
Superfamily:Upper collar protein gp10 (connector protein)
Family:Upper collar protein gp10 (connector protein) |
|
|
| 26 | c1ijgE_ |
|
not modelled |
50.5 |
15 |
PDB header:viral protein
Chain: E: PDB Molecule:upper collar protein;
PDBTitle: structure of the bacteriophage phi29 head-tail connector2 protein
|
|
|
| 27 | c5tjjA_ |
|
not modelled |
44.7 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, iclr family;
PDBTitle: crystal structure of icir transcriptional regulator from2 alicyclobacillus acidocaldarius
|
|
|
| 28 | c2kvuA_ |
|
not modelled |
37.2 |
7 |
PDB header:transcription regulator
Chain: A: PDB Molecule:mkl/myocardin-like protein 1;
PDBTitle: solution nmr structure of sap domain of mkl/myocardin-like2 protein 1 from h.sapiens, northeast structural genomics3 consortium target target hr4547e
|
|
|
| 29 | c2do1A_ |
|
not modelled |
35.6 |
12 |
PDB header:gene regulation
Chain: A: PDB Molecule:nuclear protein hcc-1;
PDBTitle: solution structure of the sap domain of human nuclear2 protein hcc-1
|
|
|
| 30 | c3k2qA_ |
|
not modelled |
35.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyrophosphate-dependent phosphofructokinase;
PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
|
|
|
| 31 | c2higA_ |
|
not modelled |
34.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:6-phospho-1-fructokinase;
PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
|
|
|
| 32 | d1k75a_ |
|
not modelled |
27.1 |
20 |
Fold:ALDH-like
Superfamily:ALDH-like
Family:L-histidinol dehydrogenase HisD |
|
|
| 33 | d1h1js_ |
|
not modelled |
26.6 |
18 |
Fold:LEM/SAP HeH motif
Superfamily:SAP domain
Family:SAP domain |
|
|
| 34 | c6an0A_ |
|
not modelled |
25.6 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histidinol dehydrogenase;
PDBTitle: crystal structure of histidinol dehydrogenase from elizabethkingia2 anophelis
|
|
|
| 35 | c3obfA_ |
|
not modelled |
23.7 |
17 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator, iclr family;
PDBTitle: crystal structure of putative transcriptional regulator, iclr family;2 targeted domain 129...302
|
|
|
| 36 | c4m1eC_ |
|
not modelled |
23.5 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase i from2 planctomyces limnophilus dsm 3776, nysgrc target 029364.
|
|
|
| 37 | c4e94A_ |
|
not modelled |
23.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:mccc family protein;
PDBTitle: crystal structure of mccf-like protein from streptococcus pneumoniae
|
|
|
| 38 | c1zrjA_ |
|
not modelled |
23.2 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:e1b-55kda-associated protein 5 isoform c;
PDBTitle: solution structure of the sap domain of human e1b-55kda-2 associated protein 5 isoform c
|
|
|
| 39 | c5xoeA_ |
|
not modelled |
23.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:atp-dependent 6-phosphofructokinase;
PDBTitle: crystal structure of the apo staphylococcus aureus phosphofructokinase
|
|
|
| 40 | d2do1a1 |
|
not modelled |
21.7 |
11 |
Fold:LEM/SAP HeH motif
Superfamily:SAP domain
Family:SAP domain |
|
|
| 41 | c1dvpA_ |
|
not modelled |
21.7 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine
PDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
|
|
|
| 42 | c3zyqA_ |
|
not modelled |
21.0 |
7 |
PDB header:signaling
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.48 a3 resolution
|
|
|
| 43 | d1zrja1 |
|
not modelled |
20.6 |
14 |
Fold:LEM/SAP HeH motif
Superfamily:SAP domain
Family:SAP domain |
|
|
| 44 | c4lnaA_ |
|
not modelled |
20.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase i from spirosoma2 linguale dsm 74, nysgrc target 029362
|
|
|
| 45 | c2c4kD_ |
|
not modelled |
20.2 |
24 |
PDB header:regulatory protein
Chain: D: PDB Molecule:phosphoribosyl pyrophosphate synthetase-associated protein
PDBTitle: crystal structure of human phosphoribosylpyrophosphate synthetase-2 associated protein 39 (pap39)
|
|
|
| 46 | c2na9A_ |
|
not modelled |
20.0 |
50 |
PDB header:signaling protein
Chain: A: PDB Molecule:cytokine receptor common subunit beta;
PDBTitle: transmembrane structure of the p441a mutant of the cytokine receptor2 common subunit beta
|
|
|
| 47 | c4g4sP_ |
|
not modelled |
19.8 |
27 |
PDB header:hydrolase/chaperone
Chain: P: PDB Molecule:proteasome assembly chaperone 2;
PDBTitle: structure of proteasome-pba1-pba2 complex
|
|
|
| 48 | c4uc0A_ |
|
not modelled |
19.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of a purine nucleoside phosphorylase (psi-nysgrc-2 029736) from agrobacterium vitis
|
|
|
| 49 | c2na8A_ |
|
not modelled |
19.3 |
50 |
PDB header:membrane protein
Chain: A: PDB Molecule:cytokine receptor common subunit beta;
PDBTitle: transmembrane structure of the cytokine receptor common subunit beta
|
|
|
| 50 | c4nsnC_ |
|
not modelled |
18.6 |
10 |
PDB header:transferase
Chain: C: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from2 porphyromonas gingivalis atcc 33277, nysgrc target 030972,3 orthorhombic symmetry
|
|
|
| 51 | d1w53a_ |
|
not modelled |
18.2 |
12 |
Fold:KaiA/RbsU domain
Superfamily:KaiA/RbsU domain
Family:Phosphoserine phosphatase RsbU, N-terminal domain |
|
|
| 52 | c4g07A_ |
|
not modelled |
18.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histidinol dehydrogenase;
PDBTitle: the crystal structure of the c366s mutant of hdh from brucella suis
|
|
|
| 53 | c4l5cE_ |
|
not modelled |
18.1 |
11 |
PDB header:transferase
Chain: E: PDB Molecule:s-methyl-5'-thioadenosine phosphorylase;
PDBTitle: methylthioadenosine phosphorylase from schistosoma mansoni in complex2 with adenine in space group p212121
|
|
|
| 54 | d2f48a1 |
|
not modelled |
16.8 |
20 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 55 | d3claa_ |
|
not modelled |
16.7 |
7 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 56 | d1li4a2 |
|
not modelled |
16.7 |
25 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:S-adenosylhomocystein hydrolase |
|
|
| 57 | d1saza2 |
|
not modelled |
16.3 |
7 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Acetokinase-like |
|
|
| 58 | c4twbB_ |
|
not modelled |
16.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: sulfolobus solfataricus ribose-phosphate pyrophosphokinase
|
|
|
| 59 | c3p0jD_ |
|
not modelled |
16.0 |
14 |
PDB header:ligase
Chain: D: PDB Molecule:tyrosyl-trna synthetase;
PDBTitle: leishmania major tyrosyl-trna synthetase in complex with tyrosinol,2 triclinic crystal form 1
|
|
|
| 60 | c2kveA_ |
|
not modelled |
15.9 |
17 |
PDB header:hormone
Chain: A: PDB Molecule:mesencephalic astrocyte-derived neurotrophic factor;
PDBTitle: c-terminal domain of mesencephalic astrocyte-derived neurotrophic2 factor (manf)
|
|
|
| 61 | c5f1yA_ |
|
not modelled |
15.9 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:mccc family protein;
PDBTitle: crystal structure of ba3275, the member of s66 family of serine2 peptidases
|
|
|
| 62 | d1y88a1 |
|
not modelled |
15.7 |
11 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:Hypothetical protein AF1548, C-terminal domain |
|
|
| 63 | c2iv3B_ |
|
not modelled |
15.6 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyltransferase;
PDBTitle: crystal structure of avigt4, a glycosyltransferase involved2 in avilamycin a biosynthesis
|
|
|
| 64 | c4gicB_ |
|
not modelled |
14.9 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histidinol dehydrogenase;
PDBTitle: crystal structure of a putative histidinol dehydrogenase (target psi-2 014034) from methylococcus capsulatus
|
|
|
| 65 | c3ilvA_ |
|
not modelled |
14.9 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:glutamine-dependent nad(+) synthetase;
PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
|
|
|
| 66 | c4grdA_ |
|
not modelled |
14.9 |
22 |
PDB header:lyase,isomerase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from burkholderia cenocepacia j2315
|
|
|
| 67 | d2dloa2 |
|
not modelled |
14.9 |
56 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
|
|
| 68 | d1q23a_ |
|
not modelled |
14.8 |
17 |
Fold:CoA-dependent acyltransferases
Superfamily:CoA-dependent acyltransferases
Family:CAT-like |
|
|
| 69 | d1zhva2 |
|
not modelled |
14.7 |
19 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
|
|
| 70 | c2kqvA_ |
|
not modelled |
14.5 |
35 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: sars coronavirus-unique domain (sud): three-domain molecular2 architecture in solution and rna binding. i: structure of the sud-m3 domain of sud-mc
|
|
|
| 71 | d1dvpa1 |
|
not modelled |
14.4 |
9 |
Fold:alpha-alpha superhelix
Superfamily:ENTH/VHS domain
Family:VHS domain |
|
|
| 72 | d4pfka_ |
|
not modelled |
14.3 |
14 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
|
|
| 73 | c2lz1A_ |
|
not modelled |
14.0 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:nuclear factor erythroid 2-related factor 2;
PDBTitle: solution nmr structure of the dna-binding domain of human nf-e2-2 related factor 2, northeast structural genomics consortium (nesg)3 target hr3520o
|
|
|
| 74 | c2vc6A_ |
|
not modelled |
13.9 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of mosa from s. meliloti with pyruvate bound
|
|
|
| 75 | d1ggaa2 |
|
not modelled |
13.4 |
23 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
| 76 | d1g99a2 |
|
not modelled |
13.2 |
3 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Acetokinase-like |
|
|
| 77 | c2jzfA_ |
|
not modelled |
12.9 |
35 |
PDB header:viral protein
Chain: A: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: nmr conformer closest to the mean coordinates of the domain 513-651 of2 the sars-cov nonstructural protein nsp3
|
|
|
| 78 | c2i9dC_ |
|
not modelled |
12.9 |
8 |
PDB header:transferase
Chain: C: PDB Molecule:chloramphenicol acetyltransferase;
PDBTitle: chloramphenicol acetyltransferase
|
|
|
| 79 | d1tf1a_ |
|
not modelled |
12.8 |
20 |
Fold:Profilin-like
Superfamily:GAF domain-like
Family:IclR ligand-binding domain-like |
|
|
| 80 | c2jmkA_ |
|
not modelled |
12.7 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:hypothetical protein ta0956;
PDBTitle: solution structure of ta0956
|
|
|
| 81 | c3dahB_ |
|
not modelled |
12.6 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:ribose-phosphate pyrophosphokinase;
PDBTitle: 2.3 a crystal structure of ribose-phosphate pyrophosphokinase from2 burkholderia pseudomallei
|
|
|
| 82 | d1sknp_ |
|
not modelled |
12.5 |
14 |
Fold:A DNA-binding domain in eukaryotic transcription factors
Superfamily:A DNA-binding domain in eukaryotic transcription factors
Family:A DNA-binding domain in eukaryotic transcription factors |
|
|
| 83 | c4qmkB_ |
|
not modelled |
12.5 |
41 |
PDB header:toxin
Chain: B: PDB Molecule:type iii secretion system effector protein exou;
PDBTitle: crystal structure of type iii effector protein exou (exou)
|
|
|
| 84 | c2g7uB_ |
|
not modelled |
12.0 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: 2.3 a structure of putative catechol degradative operon regulator from2 rhodococcus sp. rha1
|
|
|
| 85 | c6aphA_ |
|
not modelled |
11.9 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of adenosylhomocysteinase from elizabethkingia2 anophelis nuhp1 in complex with nad and adenosine
|
|
|
| 86 | c5ifkC_ |
|
not modelled |
11.8 |
25 |
PDB header:transferase
Chain: C: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: purine nucleoside phosphorylase
|
|
|
| 87 | c3mq0A_ |
|
not modelled |
11.7 |
25 |
PDB header:transcription repressor
Chain: A: PDB Molecule:transcriptional repressor of the blcabc operon;
PDBTitle: crystal structure of agobacterium tumefaciens repressor blcr
|
|
|
| 88 | c3c5yD_ |
|
not modelled |
11.4 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
|
|
| 89 | c2mfqB_ |
|
not modelled |
11.1 |
50 |
PDB header:transferase
Chain: B: PDB Molecule:bdnf/nt-3 growth factors receptor;
PDBTitle: nmr solution structures of frs2a ptb domain with neurotrophin receptor2 trkb
|
|
|
| 90 | c1zxxA_ |
|
not modelled |
11.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:6-phosphofructokinase;
PDBTitle: the crystal structure of phosphofructokinase from lactobacillus2 delbrueckii
|
|
|
| 91 | c3p4iA_ |
|
not modelled |
10.9 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:acetate kinase;
PDBTitle: crystal structure of acetate kinase from mycobacterium avium
|
|
|
| 92 | c1wtjB_ |
|
not modelled |
10.8 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ureidoglycolate dehydrogenase;
PDBTitle: crystal structure of delta1-piperideine-2-carboxylate2 reductase from pseudomonas syringae pvar.tomato
|
|
|
| 93 | c1vbiA_ |
|
not modelled |
10.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:type 2 malate/lactate dehydrogenase;
PDBTitle: crystal structure of type 2 malate/lactate dehydrogenase from thermus2 thermophilus hb8
|
|
|
| 94 | c2hkeB_ |
|
not modelled |
10.6 |
30 |
PDB header:lyase
Chain: B: PDB Molecule:diphosphomevalonate decarboxylase, putative;
PDBTitle: mevalonate diphosphate decarboxylase from trypanosoma brucei
|
|
|
| 95 | d2g82a2 |
|
not modelled |
10.6 |
22 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
| 96 | c3izbF_ |
|
not modelled |
10.6 |
16 |
PDB header:ribosome
Chain: F: PDB Molecule:40s ribosomal protein rps5 (s7p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 97 | c3dezA_ |
|
not modelled |
10.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:orotate phosphoribosyltransferase;
PDBTitle: crystal structure of orotate phosphoribosyltransferase from2 streptococcus mutans
|
|
|
| 98 | d1i27a_ |
|
not modelled |
10.5 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal domain of the rap74 subunit of TFIIF |
|
|
| 99 | d1v8ba2 |
|
not modelled |
10.1 |
25 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:S-adenosylhomocystein hydrolase |
|
|