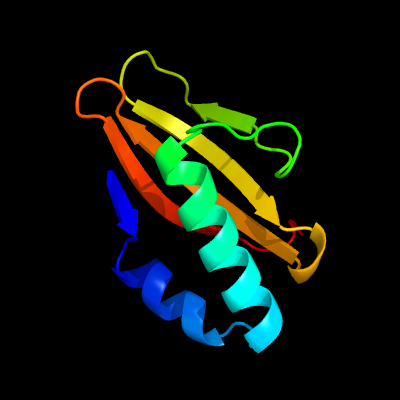
| 1 | d1wmia1
|
|
 |
98.7 |
18 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
|
|
|
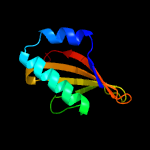
| 2 | c2kheA_
|
|
 |
98.5 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxin-like protein;
PDBTitle: solution structure of the bacterial toxin rele from thermus2 thermophilus hb8
|
|
|
|
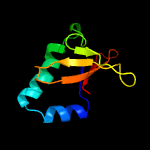
| 3 | c3bpqD_
|
|
 |
98.5 |
9 |
PDB header:toxin
Chain: D: PDB Molecule:toxin rele3;
PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
|
|
|
|
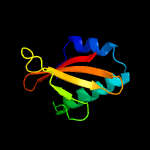
| 4 | c3kixy_
|
|
 |
98.2 |
10 |
PDB header:ribosome
Chain: Y: PDB Molecule:
PDBTitle: structure of rele nuclease bound to the 70s ribosome (postcleavage2 state; part 3 of 4)
|
|
|
|
| 5 | c3g5oC_
|
|
 |
98.0 |
19 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
|
|
|
| 6 | d2a6sa1
|
|
 |
97.7 |
10 |
Fold:RelE-like
Superfamily:RelE-like
Family:YoeB/Txe-like |
|
|
|
| 7 | c3oeiH_
|
|
 |
97.5 |
15 |
PDB header:toxin, protein binding
Chain: H: PDB Molecule:relk (toxin rv3358);
PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
|
|
|
|
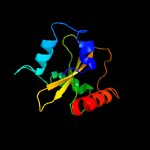
| 8 | c5ja9D_
|
|
 |
96.8 |
21 |
PDB header:toxin
Chain: D: PDB Molecule:toxin higb-2;
PDBTitle: crystal structure of the higb2 toxin in complex with nb6
|
|
|
|
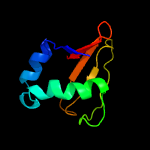
| 9 | c2otrA_
|
|
 |
95.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein hp0892;
PDBTitle: solution structure of conserved hypothetical protein hp0892 from2 helicobacter pylori
|
|
|
|
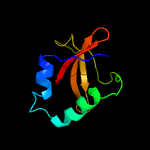
| 10 | d1z8ma1
|
|
 |
90.8 |
20 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
|
|
|
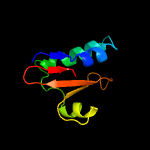
| 11 | c5cw7H_
|
|
 |
84.6 |
16 |
PDB header:toxin
Chain: H: PDB Molecule:plasmid stabilization protein pare;
PDBTitle: crystal structure of the paaa2-pare2 antitoxin-toxin complex
|
|
|
|
| 12 | c4q2uH_
|
|
 |
83.3 |
8 |
PDB header:toxin/toxin repressor
Chain: H: PDB Molecule:mrna interferase yafq;
PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
|
|
|
|
| 13 | c4mctD_
|
|
 |
77.8 |
10 |
PDB header:toxin
Chain: D: PDB Molecule:killer protein;
PDBTitle: p. vulgaris higba structure, crystal form 1
|
|
|
|
| 14 | c5cegB_
|
|
 |
47.9 |
11 |
PDB header:toxin
Chain: B: PDB Molecule:plasmid stabilization system;
PDBTitle: x-ray structure of toxin/anti-toxin complex from mesorhizobium2 opportunistum
|
|
|
|
| 15 | c6f8sD_
|
|
 |
18.1 |
11 |
PDB header:toxin
Chain: D: PDB Molecule:putative killer protein;
PDBTitle: toxin-antitoxin complex grata
|
|
|
|
| 16 | c2kl4A_
|
|
 |
17.3 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh2032 protein;
PDBTitle: nmr structure of the protein nb7804a
|
|
|
|
| 17 | c2ogfD_
|
|
 |
15.3 |
26 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein mj0408;
PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
|
|
|
|
| 18 | d2oc6a1
|
|
 |
12.4 |
10 |
Fold:Secretion chaperone-like
Superfamily:YdhG-like
Family:YdhG-like |
|
|
|
| 19 | d1kfia4
|
|
 |
8.6 |
32 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
|
| 20 | c3kxeB_
|
|
 |
7.9 |
15 |
PDB header:protein binding
Chain: B: PDB Molecule:toxin protein pare-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
|
|
|
| 21 | c4jenB_ |
|
not modelled |
7.9 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:cmp n-glycosidase;
PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
|
|
|
| 22 | c3bdiA_ |
|
not modelled |
7.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ta0194;
PDBTitle: crystal structure of predicted cib-like hydrolase (np_393672.1) from2 thermoplasma acidophilum at 1.45 a resolution
|
|
|
| 23 | c3kxeD_ |
|
not modelled |
7.1 |
13 |
PDB header:protein binding
Chain: D: PDB Molecule:antitoxin protein pard-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
|
|
| 24 | c5cegC_ |
|
not modelled |
7.0 |
19 |
PDB header:toxin
Chain: C: PDB Molecule:addiction module antidote protein, copg/arc/metj family;
PDBTitle: x-ray structure of toxin/anti-toxin complex from mesorhizobium2 opportunistum
|
|
|
| 25 | c5frdA_ |
|
not modelled |
6.5 |
5 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxylesterase (est-2);
PDBTitle: structure of a thermophilic esterase
|
|
|
| 26 | c4qwoA_ |
|
not modelled |
6.0 |
13 |
PDB header:viral protein
Chain: A: PDB Molecule:profilin;
PDBTitle: 1.52 angstrom crystal structure of a42r profilin-like protein from2 monkeypox virus zaire-96-i-16
|
|
|
| 27 | d1eiwa_ |
|
not modelled |
6.0 |
16 |
Fold:Flavodoxin-like
Superfamily:Hypothetical protein MTH538
Family:Hypothetical protein MTH538 |
|
|
| 28 | c4lgdF_ |
|
not modelled |
5.9 |
10 |
PDB header:signaling protein
Chain: F: PDB Molecule:ras association domain family member 5, rassf5;
PDBTitle: structural basis for autoactivation of human mst2 kinase and its2 regulation by rassf5
|
|
|
| 29 | c1u2eA_ |
|
not modelled |
5.9 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:2-hydroxy-6-ketonona-2,4-dienedioic acid
PDBTitle: crystal structure of the c-c bond hydrolase mhpc
|
|
|
| 30 | c2fu4B_ |
|
not modelled |
5.8 |
18 |
PDB header:dna binding protein
Chain: B: PDB Molecule:ferric uptake regulation protein;
PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
|
|
|
| 31 | c1fi4A_ |
|
not modelled |
5.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:mevalonate 5-diphosphate decarboxylase;
PDBTitle: the x-ray crystal structure of mevalonate 5-diphosphate decarboxylase2 at 2.3 angstrom resolution.
|
|
|
| 32 | d3pmga4 |
|
not modelled |
5.6 |
18 |
Fold:TBP-like
Superfamily:Phosphoglucomutase, C-terminal domain
Family:Phosphoglucomutase, C-terminal domain |
|
|
| 33 | c2p90B_ |
|
not modelled |
5.5 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein cgl1923;
PDBTitle: the crystal structure of a protein of unknown function from2 corynebacterium glutamicum atcc 13032
|
|
|
| 34 | c1psvA_ |
|
not modelled |
5.4 |
42 |
PDB header:designed peptide
Chain: A: PDB Molecule:pda8d;
PDBTitle: computationally designed peptide with a beta-beta-alpha2 fold selection, nmr, 32 structures
|
|
|
| 35 | d1ss4a_ |
|
not modelled |
5.3 |
19 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Hypothetical protein BC1747 |
|
|