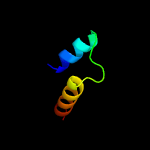
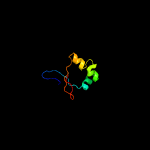
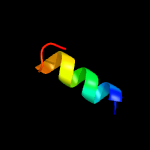
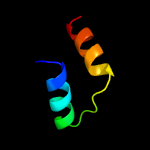
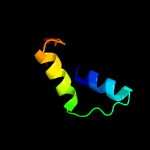
1 c5fimA_
24.7
14
PDB header: unknown functionChain: A: PDB Molecule: ygau;PDBTitle: the structure of kbp.k from e. coli
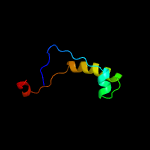
2 d1y7ma2
24.5
20
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain
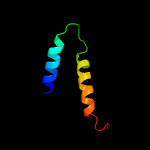
3 c2mkxA_
21.2
30
PDB header: hydrolaseChain: A: PDB Molecule: autolysin;PDBTitle: solution structure of lysm the peptidoglycan binding domain of2 autolysin atla from enterococcus faecalis
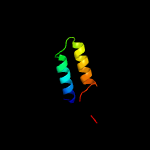
4 c1ks9A_
20.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 2-dehydropantoate 2-reductase;PDBTitle: ketopantoate reductase from escherichia coli
5 d1ks9a1
18.7
19
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Ketopantoate reductase PanE
6 d1e0ga_
16.3
40
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain
7 c4pqlB_
14.5
20
PDB header: dna binding proteinChain: B: PDB Molecule: truncated replication protein repa;PDBTitle: n-terminal domain of dna binding protein
8 c2yonA_
14.3
50
PDB header: signaling proteinChain: A: PDB Molecule: sensory box protein;PDBTitle: solution nmr structure of the c-terminal extension of two bacterial2 light, oxygen, voltage (lov) photoreceptor proteins from3 pseudomonas putida
9 c3no7A_
13.8
53
PDB header: dna binding proteinChain: A: PDB Molecule: putative plasmid related protein;PDBTitle: crystal structure of the centromere-binding protein parb from plasmid2 pcxc100
10 c5zhcA_
12.4
30
PDB header: dna binding proteinChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of the padr-family transcriptional regulator rv34882 of mycobacterium tuberculosis h37rv
11 d2ev0a1
12.4
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein
12 c3k96B_
12.3
33
PDB header: oxidoreductaseChain: B: PDB Molecule: glycerol-3-phosphate dehydrogenase [nad(p)+];PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
13 c5ocdD_
12.0
29
PDB header: rna binding proteinChain: D: PDB Molecule: cyclodipeptide synthase;PDBTitle: structure of a cdps from fluoribacter dumoffii
14 c5nbcD_
10.6
22
PDB header: dna binding proteinChain: D: PDB Molecule: ferric uptake regulation protein;PDBTitle: structure of prokaryotic transcription factors
15 c2djpA_
10.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein sb145;PDBTitle: the solution structure of the lysm domain of human2 hypothetical protein sb145
16 d2nrac1
10.2
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Replication initiation protein
17 d1fipa_
10.1
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like
18 c4b3nA_
10.1
29
PDB header: sugar binding protein/ligaseChain: A: PDB Molecule: maltose-binding periplasmic protein, tripartite motif-PDBTitle: crystal structure of rhesus trim5alpha pry/spry domain
19 c4pxvC_
10.1
20
PDB header: sugar binding proteinChain: C: PDB Molecule: chitinase a;PDBTitle: crystal structure of lysm domain from pteris ryukyuensis chitinase a
20 c6hu9e_
9.8
23
PDB header: oxidoreductase/electron transportChain: E: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
21 d1g3wa1
not modelled
9.7
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein
22 d1dj0a_
not modelled
9.6
21
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: Pseudouridine synthase I TruA
23 c5k2lA_
not modelled
9.3
50
PDB header: hydrolaseChain: A: PDB Molecule: chitinase, lysozyme;PDBTitle: crystal structure of lysm domain from volvox carteri chitinase
24 c5l9wb_
not modelled
9.2
58
PDB header: ligaseChain: B: PDB Molecule: acetophenone carboxylase gamma subunit;PDBTitle: crystal structure of the apc core complex
25 d2itba1
not modelled
9.2
19
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: MiaE-like
26 c5bumA_
not modelled
8.5
40
PDB header: sugar binding proteinChain: A: PDB Molecule: chitinase a;PDBTitle: crystal structure of lysm domain from equisetum arvense chitinase a
27 c3bjqA_
not modelled
8.4
35
PDB header: viral proteinChain: A: PDB Molecule: phage-related protein;PDBTitle: crystal structure of a phage-related protein (bb3626) from bordetella2 bronchiseptica rb50 at 2.05 a resolution
28 c5l9wB_
not modelled
8.4
67
PDB header: ligaseChain: B: PDB Molecule: acetophenone carboxylase gamma subunit;PDBTitle: crystal structure of the apc core complex
29 d2isya1
not modelled
7.8
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein
30 d1q6za2
not modelled
7.8
31
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Pyruvate oxidase and decarboxylase Pyr module
31 d1v9pa3
not modelled
7.7
25
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase
32 d1mzba_
not modelled
7.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like
33 c1jpxA_
not modelled
7.1
24
PDB header: viral proteinChain: A: PDB Molecule: gp41 envelope protein;PDBTitle: mutation that destabilize the gp41 core: determinants for stabilizing2 the siv/cpmac envelope glycoprotein complex. wild type.
34 c4p96B_
not modelled
6.9
21
PDB header: transcriptionChain: B: PDB Molecule: fatty acid metabolism regulator protein;PDBTitle: fadr, fatty acid responsive transcription factor from vibrio cholerae
35 c2l9yA_
not modelled
6.4
30
PDB header: sugar binding proteinChain: A: PDB Molecule: cvnh-lysm lectin;PDBTitle: solution structure of the mocvnh-lysm module from the rice blast2 fungus magnaporthe oryzae protein (mgg_03307)
36 c1hsjA_
not modelled
6.2
26
PDB header: transcription/sugar binding proteinChain: A: PDB Molecule: fusion protein consisting of staphylococcus accessoryPDBTitle: sarr mbp fusion structure
37 d3lada3
not modelled
6.1
27
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
38 c5fgmA_
not modelled
5.8
19
PDB header: hydrolaseChain: A: PDB Molecule: ecf rna polymerase sigma factor sigr;PDBTitle: streptomyces coelicolor sigr region 4
39 d1w7pd1
not modelled
5.8
30
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain
40 c3g17H_
not modelled
5.7
17
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: similar to 2-dehydropantoate 2-reductase;PDBTitle: structure of putative 2-dehydropantoate 2-reductase from2 staphylococcus aureus
41 c6dlcB_
not modelled
5.7
43
PDB header: de novo proteinChain: B: PDB Molecule: designed protein dhd1:234_b;PDBTitle: designed protein dhd1:234_a, designed protein dhd1:234_b
42 c4bryB_
not modelled
5.7
35
PDB header: cell cycleChain: B: PDB Molecule: multicilin;PDBTitle: the idas:geminin heterodimeric parallel coiled-coil
43 c2kelB_
not modelled
5.6
50
PDB header: transcription repressorChain: B: PDB Molecule: uncharacterized protein 56b;PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
44 c4uz2D_
not modelled
5.4
30
PDB header: hydrolaseChain: D: PDB Molecule: cell wall-binding endopeptidase-related protein;PDBTitle: crystal structure of the n-terminal lysm domains from the putative2 nlpc/p60 d,l endopeptidase from t. thermophilus
45 c3f10A_
not modelled
5.4
8
PDB header: hydrolase, lyaseChain: A: PDB Molecule: 8-oxoguanine-dna-glycosylase;PDBTitle: crystal structure of clostridium acetobutylicum 8-oxoguanine dna2 glycosylase in complex with 8-oxoguanosine
46 c3mwmA_
not modelled
5.3
17
PDB header: transcriptionChain: A: PDB Molecule: putative metal uptake regulation protein;PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur