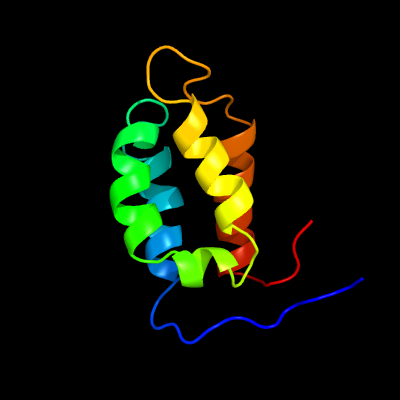
1 c5frhA_
99.7
22
PDB header: transcriptionChain: A: PDB Molecule: anti-sigma factor rsra;PDBTitle: solution structure of oxidised rsra
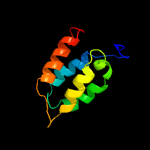
2 c5wuqD_
99.4
26
PDB header: metal binding proteinChain: D: PDB Molecule: anti-sigma-w factor rsiw;PDBTitle: crystal structure of sigw in complex with its anti-sigma rsiw, a zinc2 binding form
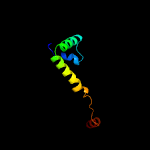
3 c3vdoB_
99.3
13
PDB header: dna binding protein/protein bindingChain: B: PDB Molecule: anti-sigma-k factor rska;PDBTitle: structure of extra-cytoplasmic function(ecf) sigma factor sigk in2 complex with its negative regulator rska from mycobacterium3 tuberculosis
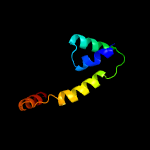
4 c2z2sD_
98.9
14
PDB header: transcriptionChain: D: PDB Molecule: anti-sigma factor chrr, transcriptional activator chrr;PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
5 c3hugJ_
98.9
20
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
6 c6in7A_
98.9
6
PDB header: transcriptionChain: A: PDB Molecule: sigma factor algu negative regulatory protein;PDBTitle: crystal structure of algu in complex with muca(cyto)
7 d1or7c_
97.2
18
Fold: N-terminal, cytoplasmic domain of anti-sigmaE factor RseASuperfamily: N-terminal, cytoplasmic domain of anti-sigmaE factor RseAFamily: N-terminal, cytoplasmic domain of anti-sigmaE factor RseA
8 c1or7C_
97.2
18
PDB header: transcriptionChain: C: PDB Molecule: sigma-e factor negative regulatory protein;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
9 c5camC_
40.6
8
PDB header: transcriptionChain: C: PDB Molecule: pupr protein;PDBTitle: crystal structure of the cytoplasmic domain of the pseudomonas putida2 anti-sigma factor pupr (semet)
10 c2xuvB_
40.1
27
PDB header: unknown functionChain: B: PDB Molecule: hdeb;PDBTitle: the structure of hdeb
11 d2db7a1
39.3
27
Fold: Orange domain-likeSuperfamily: Orange domain-likeFamily: Hairy Orange domain
12 c5fmnB_
33.1
29
PDB header: dna binding proteinChain: B: PDB Molecule: inrs;PDBTitle: the nickel-responsive transcriptional regulator inrs
13 c5ze4A_
28.9
21
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase, chloroplastic;PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
14 c2hh7A_
27.7
26
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein csor;PDBTitle: crystal structure of cu(i) bound csor from mycobacterium tuberculosis.
15 c5lcyD_
27.3
16
PDB header: transcriptionChain: D: PDB Molecule: frmr;PDBTitle: formaldehyde-responsive regulator frmr e64h variant from salmonella2 enterica serovar typhimurium
16 c3aaiB_
26.2
23
PDB header: transcriptionChain: B: PDB Molecule: copper homeostasis operon regulatory protein;PDBTitle: x-ray crystal structure of csor from thermus thermophilus hb8
17 c5lbmD_
24.5
16
PDB header: transcriptionChain: D: PDB Molecule: transcriptional repressor frmr;PDBTitle: the asymmetric tetrameric structure of the formaldehyde sensing2 transcriptional repressor frmr from escherichia coli
18 c4m1pA_
23.7
16
PDB header: transcription repressorChain: A: PDB Molecule: copper-sensitive operon repressor (csor);PDBTitle: crystal structure of the copper-sensing repressor csor with cu(i) from2 geobacillus thermodenitrificans ng80-2
19 c4adzA_
23.1
19
PDB header: transcriptionChain: A: PDB Molecule: csor;PDBTitle: crystal structure of the apo form of a copper-sensitive operon2 regulator (csor) protein from streptomyces lividans
20 c5oynB_
22.9
14
PDB header: lyaseChain: B: PDB Molecule: dehydratase, ilvd/edd family;PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
21 c4xvvB_
not modelled
22.9
13
PDB header: chaperoneChain: B: PDB Molecule: acid stress chaperone hdeb;PDBTitle: crystal structure of an acid stress chaperone hdeb (kpn_03484) from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.70 a3 resolution
22 c2lkyA_
not modelled
22.2
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
23 c5x56A_
not modelled
16.8
10
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii repair protein psb27-h1, chloroplastic;PDBTitle: crystal structure of psb27 from arabidopsis thaliana
24 d2elca1
not modelled
16.3
25
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
25 c5j84A_
not modelled
15.8
7
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase;PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
26 d1khda1
not modelled
15.4
8
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
27 d1uoua1
not modelled
15.2
13
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
28 c4jd9B_
not modelled
15.2
20
PDB header: protein bindingChain: B: PDB Molecule: 14.5 kda salivary protein;PDBTitle: contact pathway inhibitor from a sand fly
29 c2kvcA_
not modelled
15.2
20
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
30 c3mayE_
not modelled
14.0
17
PDB header: heme-binding proteinChain: E: PDB Molecule: possible exported protein;PDBTitle: crystal structure of a secreted mycobacterium tuberculosis heme-2 binding protein
31 c6gvkB_
not modelled
14.0
80
PDB header: structural proteinChain: B: PDB Molecule: dystonin;PDBTitle: second pair of fibronectin type iii domains of integrin beta4 (t1663r2 mutant) bound to the bullous pemphigoid antigen bp230 (bpag1e)
32 c6gvlB_
not modelled
13.9
80
PDB header: structural proteinChain: B: PDB Molecule: dystonin;PDBTitle: second pair of fibronectin type iii domains of integrin beta4 bound to2 the bullous pemphigoid antigen bp230 (bpag1e)
33 c2yreA_
not modelled
13.3
50
PDB header: protein bindingChain: A: PDB Molecule: f-box only protein 30;PDBTitle: solution structure of the zinc finger domains (1-87) from2 human f-box only protein
34 d2pp4a1
not modelled
12.7
26
Fold: TAFH domain-likeSuperfamily: TAFH domain-likeFamily: TAFH domain-like
35 d2hwna1
not modelled
12.6
29
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
36 c2wfbA_
not modelled
11.7
33
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
37 c5edvA_
not modelled
11.4
17
PDB header: ligase/transferaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase rnf31;PDBTitle: structure of the hoip-rbr/ubch5b~ubiquitin transfer complex
38 c3jb9i_
not modelled
11.3
15
PDB header: rna binding protein/rnaChain: I: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
39 d1k8ia2
not modelled
11.0
29
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain
40 c5m45I_
not modelled
10.7
13
PDB header: ligaseChain: I: PDB Molecule: acetone carboxylase gamma subunit;PDBTitle: structure of acetone carboxylase purified from xanthobacter2 autotrophicus
41 c2lm4A_
not modelled
10.7
13
PDB header: protein bindingChain: A: PDB Molecule: succinate dehydrogenase assembly factor 2, mitochondrial;PDBTitle: solution nmr structure of mitochondrial succinate dehydrogenase2 assembly factor 2 from saccharomyces cerevisiae, northeast structural3 genomics consortium target yt682a
42 d1wjva1
not modelled
10.7
25
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: C2HC finger
43 d1puza_
not modelled
10.4
11
Fold: YgfY-likeSuperfamily: YgfY-likeFamily: YgfY-like
44 c2kmfA_
not modelled
10.3
17
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kda protein;PDBTitle: solution structure of psb27 from cyanobacterial photosystem2 ii
45 d1r2aa_
not modelled
9.9
23
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
46 d1k5oa_
not modelled
9.7
24
Fold: Myosin phosphatase inhibitor 17kDa protein, CPI-17Superfamily: Myosin phosphatase inhibitor 17kDa protein, CPI-17Family: Myosin phosphatase inhibitor 17kDa protein, CPI-17
47 d1o17a1
not modelled
9.2
14
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
48 d2b5ea3
not modelled
9.2
24
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: PDI-like
49 d1v8ga1
not modelled
9.1
22
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
50 d1j2na_
not modelled
9.0
24
Fold: Myosin phosphatase inhibitor 17kDa protein, CPI-17Superfamily: Myosin phosphatase inhibitor 17kDa protein, CPI-17Family: Myosin phosphatase inhibitor 17kDa protein, CPI-17
51 d1jroa1
not modelled
8.7
23
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
52 d2tpta1
not modelled
8.6
7
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
53 d1ffva1
not modelled
8.6
15
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
54 d1n62a1
not modelled
8.4
20
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
55 c2qtdA_
not modelled
7.9
20
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein mj0327;PDBTitle: crystal structure of a putative dinitrogenase (mj0327) from2 methanocaldococcus jannaschii dsm at 1.70 a resolution
56 c2re2A_
not modelled
7.9
27
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized protein ta1041;PDBTitle: crystal structure of a putative iron-molybdenum cofactor (femo-co)2 dinitrogenase (ta1041m) from thermoplasma acidophilum dsm 1728 at3 1.30 a resolution
57 d1brwa1
not modelled
7.6
14
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
58 c2y6xA_
not modelled
7.4
16
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kd protein;PDBTitle: structure of psb27 from thermosynechococcus elongatus
59 c3b73A_
not modelled
7.1
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phih1 repressor-like protein;PDBTitle: crystal structure of the phih1 repressor-like protein from haloarcula2 marismortui
60 c2clbP_
not modelled
7.0
12
PDB header: metal binding proteinChain: P: PDB Molecule: dps-like protein;PDBTitle: the structure of the dps-like protein from sulfolobus2 solfataricus reveals a bacterioferritin-like di-metal3 binding site within a dps-like dodecameric assembly
61 d2f8aa1
not modelled
7.0
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
62 c2zxkB_
not modelled
6.7
22
PDB header: oxidoreductaseChain: B: PDB Molecule: red chlorophyll catabolite reductase,PDBTitle: crystal structure of semet-red chlorophyll catabolite2 reductase
63 c5d6sB_
not modelled
6.7
28
PDB header: oxidoreductaseChain: B: PDB Molecule: epoxyqueuosine reductase;PDBTitle: structure of epoxyqueuosine reductase from streptococcus thermophilus.
64 c4a1qB_
not modelled
6.6
19
PDB header: viral proteinChain: B: PDB Molecule: orf e73;PDBTitle: solution structure of e73 protein from sulfolobus spindle-2 shaped virus ragged hills, a hyperthermophilic3 crenarchaeal virus from yellowstone national park
65 c4aaiB_
not modelled
6.6
19
PDB header: viral proteinChain: B: PDB Molecule: orf e73;PDBTitle: thermostable protein from hyperthermophilic virus ssv-rh
66 d1t3va_
not modelled
6.5
27
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
67 d1e0ea_
not modelled
6.5
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: N-terminal Zn binding domain of HIV integraseFamily: N-terminal Zn binding domain of HIV integrase
68 d1v58a1
not modelled
6.5
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like
69 c2rhfA_
not modelled
6.4
31
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase recq;PDBTitle: d. radiodurans recq hrdc domain 3
70 c3ol4B_
not modelled
6.4
16
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
71 c1nohB_
not modelled
6.0
15
PDB header: viral proteinChain: B: PDB Molecule: head morphogenesis protein;PDBTitle: the structure of bacteriophage phi29 scaffolding protein2 gp7 after prohead assembly
72 d1t3ba1
not modelled
5.6
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: DsbC/DsbG C-terminal domain-like
73 c3gv1A_
not modelled
5.4
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: disulfide interchange protein;PDBTitle: crystal structure of disulfide interchange protein from neisseria2 gonorrhoeae
74 d2di0a1
not modelled
5.3
12
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: CUE domain
75 c4tr3A_
not modelled
5.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: type iii iodothyronine deiodinase;PDBTitle: mouse iodothyronine deiodinase 3 catalytic core, semet-labeled active2 site mutant secys->cys
76 c2jxuA_
not modelled
5.2
7
PDB header: unknown functionChain: A: PDB Molecule: terb;PDBTitle: nmr solution structure of kp-terb, a tellurite resistance2 protein from klebsiella pneumoniae
77 c4gtnA_
not modelled
5.2
15
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: structure of anthranilate phosphoribosyl transferase from2 acinetobacter baylyi
78 d1rdua_
not modelled
5.2
13
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
79 d2ou3a1
not modelled
5.2
19
Fold: TerB-likeSuperfamily: TerB-likeFamily: COG3793-like
80 d1k6ya1
not modelled
5.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: N-terminal Zn binding domain of HIV integraseFamily: N-terminal Zn binding domain of HIV integrase
81 d1wjfa_
not modelled
5.1
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: N-terminal Zn binding domain of HIV integraseFamily: N-terminal Zn binding domain of HIV integrase
82 c2yucA_
not modelled
5.1
33
PDB header: signaling proteinChain: A: PDB Molecule: tnf receptor-associated factor 4;PDBTitle: solution structure of the traf-type zinc finger domains2 (102-164) from human tnf receptor associated factor 4
83 c5edvB_
not modelled
5.1
17
PDB header: ligase/transferaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase rnf31;PDBTitle: structure of the hoip-rbr/ubch5b~ubiquitin transfer complex