| 1 |
|
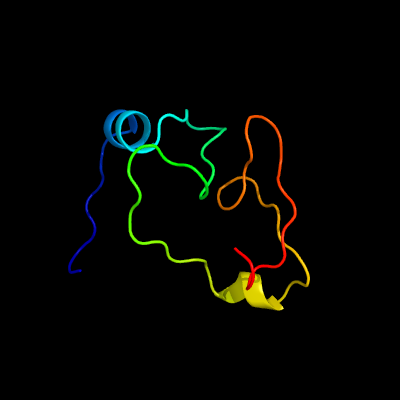
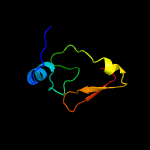
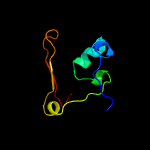
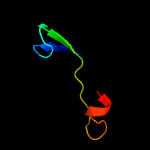
PDB 1dbx chain A
Region: 1 - 66
Aligned: 62
Modelled: 66
Confidence: 99.6%
Identity: 42%
Fold: YbaK/ProRS associated domain
Superfamily: YbaK/ProRS associated domain
Family: YbaK/ProRS associated domain
Phyre2
| 2 |
|
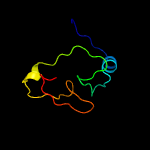
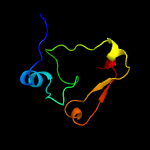
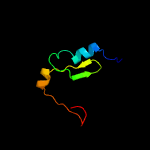
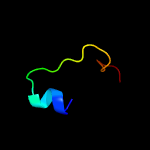
PDB 1wdv chain A
Region: 2 - 66
Aligned: 63
Modelled: 63
Confidence: 99.6%
Identity: 25%
Fold: YbaK/ProRS associated domain
Superfamily: YbaK/ProRS associated domain
Family: YbaK/ProRS associated domain
Phyre2
| 3 |
|
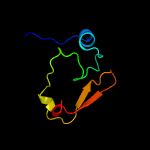
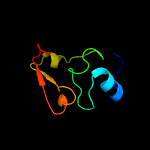
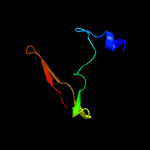
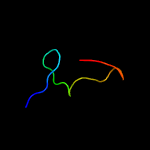
PDB 2dxa chain A
Region: 1 - 66
Aligned: 64
Modelled: 66
Confidence: 99.5%
Identity: 34%
PDB header:translation
Chain: A: PDB Molecule:protein ybak;
PDBTitle: crystal structure of trans editing enzyme prox from e.coli
Phyre2
| 4 |
|
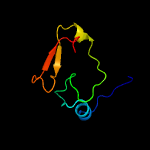
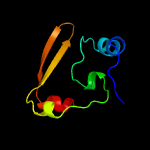
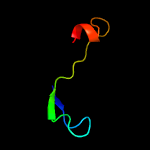
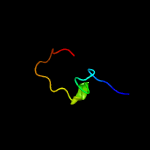
PDB 2cx5 chain B
Region: 2 - 66
Aligned: 63
Modelled: 63
Confidence: 99.5%
Identity: 27%
PDB header:translation
Chain: B: PDB Molecule:a putative trans-editing enzyme;
PDBTitle: crystal structure of a putative trans-editing enzyme for2 prolyl trna synthetase
Phyre2
| 5 |
|
PDB 1vki chain A
Region: 1 - 66
Aligned: 64
Modelled: 66
Confidence: 99.5%
Identity: 17%
Fold: YbaK/ProRS associated domain
Superfamily: YbaK/ProRS associated domain
Family: YbaK/ProRS associated domain
Phyre2
| 6 |
|
PDB 1vjf chain A
Region: 1 - 66
Aligned: 64
Modelled: 66
Confidence: 99.4%
Identity: 19%
Fold: YbaK/ProRS associated domain
Superfamily: YbaK/ProRS associated domain
Family: YbaK/ProRS associated domain
Phyre2
| 7 |
|
PDB 3mem chain A
Region: 2 - 70
Aligned: 67
Modelled: 69
Confidence: 99.4%
Identity: 10%
PDB header:signaling protein
Chain: A: PDB Molecule:putative signal transduction protein;
PDBTitle: crystal structure of a putative signal transduction protein2 (maqu_0641) from marinobacter aquaeolei vt8 at 2.25 a resolution
Phyre2
| 8 |
|
PDB 3op6 chain B
Region: 2 - 66
Aligned: 63
Modelled: 65
Confidence: 99.3%
Identity: 17%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an oligo-nucleotide binding protein (lpg1207)2 from legionella pneumophila subsp. pneumophila str. philadelphia 1 at3 2.00 a resolution
Phyre2
| 9 |
|
PDB 4d2i chain B
Region: 2 - 56
Aligned: 52
Modelled: 55
Confidence: 50.0%
Identity: 15%
PDB header:hydrolase
Chain: B: PDB Molecule:hera;
PDBTitle: crystal structure of the hera hexameric dna translocase2 from sulfolobus solfataricus bound to amp-pnp
Phyre2
| 10 |
|
PDB 3uep chain B
Region: 12 - 61
Aligned: 46
Modelled: 50
Confidence: 8.6%
Identity: 17%
PDB header:protein transport
Chain: B: PDB Molecule:yscq-c, type iii secretion protein;
PDBTitle: crystal structure of yscq-c from yersinia pseudotuberculosis
Phyre2
| 11 |
|
PDB 1xi8 chain A domain 3
Region: 36 - 63
Aligned: 28
Modelled: 28
Confidence: 8.6%
Identity: 25%
Fold: Molybdenum cofactor biosynthesis proteins
Superfamily: Molybdenum cofactor biosynthesis proteins
Family: MoeA central domain-like
Phyre2
| 12 |
|
PDB 5jpq chain D
Region: 26 - 41
Aligned: 16
Modelled: 16
Confidence: 8.3%
Identity: 19%
PDB header:ribosome
Chain: D: PDB Molecule:wd40 domain proteins;
PDBTitle: cryo-em structure of the 90s pre-ribosome
Phyre2
| 13 |
|
PDB 3bqh chain A
Region: 16 - 47
Aligned: 31
Modelled: 32
Confidence: 7.0%
Identity: 32%
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb;
PDBTitle: structure of the central domain (msra) of neisseria meningitidis pilb2 (oxidized form)
Phyre2
| 14 |
|
PDB 2j89 chain A
Region: 16 - 47
Aligned: 31
Modelled: 32
Confidence: 6.8%
Identity: 26%
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine sulfoxide reductase a;
PDBTitle: functional and structural aspects of poplar cytosolic and2 plastidial type a methionine sulfoxide reductases
Phyre2
| 15 |
|
PDB 2nqr chain A domain 3
Region: 36 - 57
Aligned: 22
Modelled: 22
Confidence: 6.4%
Identity: 14%
Fold: Molybdenum cofactor biosynthesis proteins
Superfamily: Molybdenum cofactor biosynthesis proteins
Family: MoeA central domain-like
Phyre2
| 16 |
|
PDB 2hbp chain A
Region: 1 - 17
Aligned: 17
Modelled: 17
Confidence: 6.4%
Identity: 29%
PDB header:endocytosis, protein binding
Chain: A: PDB Molecule:cytoskeleton assembly control protein sla1;
PDBTitle: solution structure of sla1 homology domain 1
Phyre2
| 17 |
|
PDB 4mj7 chain B
Region: 26 - 41
Aligned: 16
Modelled: 16
Confidence: 6.2%
Identity: 25%
PDB header:rna binding protein
Chain: B: PDB Molecule:rrna-processing protein utp23;
PDBTitle: crystal structure of the pin domain of saccharomyces cerevisiae utp23
Phyre2
| 18 |
|
PDB 1spi chain A
Region: 48 - 70
Aligned: 22
Modelled: 23
Confidence: 6.1%
Identity: 14%
Fold: Carbohydrate phosphatase
Superfamily: Carbohydrate phosphatase
Family: Inositol monophosphatase/fructose-1,6-bisphosphatase-like
Phyre2
| 19 |
|
PDB 5oez chain A
Region: 48 - 70
Aligned: 22
Modelled: 23
Confidence: 5.7%
Identity: 14%
PDB header:hydrolase
Chain: A: PDB Molecule:fbp protein;
PDBTitle: crystal structure of leishmania major fructose-1,6-bisphosphatase in2 apo form.
Phyre2
| 20 |
|
PDB 1bk4 chain A
Region: 48 - 70
Aligned: 22
Modelled: 23
Confidence: 5.5%
Identity: 18%
Fold: Carbohydrate phosphatase
Superfamily: Carbohydrate phosphatase
Family: Inositol monophosphatase/fructose-1,6-bisphosphatase-like
Phyre2
| 21 |
|
| 22 |
|