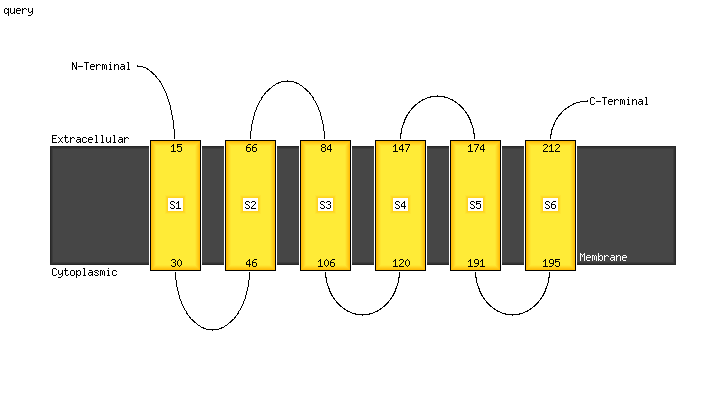
| 1 | c4a2nB_
|
|
 |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:isoprenylcysteine carboxyl methyltransferase;
PDBTitle: crystal structure of ma-icmt
|
|
|
|
| 2 | c5v7pA_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:protein-s-isoprenylcysteine o-methyltransferase;
PDBTitle: atomic structure of the eukaryotic intramembrane ras methyltransferase2 icmt (isoprenylcysteine carboxyl methyltransferase), in complex with3 a monobody
|
|
|
|
| 3 | c4quvB_
|
|
 |
99.9 |
12 |
PDB header:oxidoreductase, membrane protein
Chain: B: PDB Molecule:delta(14)-sterol reductase;
PDBTitle: structure of an integral membrane delta(14)-sterol reductase
|
|
|
|
| 4 | c4y9iA_
|
|
 |
57.6 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mycobacterium tuberculosis paralogous family 11;
PDBTitle: structure of f420-h2 dependent reductase (fdr-a) msmeg_2027
|
|
|
|
| 5 | c3r5yC_
|
|
 |
37.7 |
23 |
PDB header:unknown function
Chain: C: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of a deazaflavin-dependent nitroreductase from nocardia2 farcinica, with co-factor f420
|
|
|
|
| 6 | d1kf6c_
|
|
 |
36.2 |
17 |
Fold:Heme-binding four-helical bundle
Superfamily:Fumarate reductase respiratory complex transmembrane subunits
Family:Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD) |
|
|
|
| 7 | c3r5wO_
|
|
 |
25.6 |
23 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:deazaflavin-dependent nitroreductase;
PDBTitle: structure of ddn, the deazaflavin-dependent nitroreductase from2 mycobacterium tuberculosis involved in bioreductive activation of pa-3 824, with co-factor f420
|
|
|
|
| 8 | c5e8jC_
|
|
 |
23.8 |
20 |
PDB header:translation
Chain: C: PDB Molecule:rnmt-activating mini protein;
PDBTitle: crystal structure of mrna cap guanine-n7 methyltransferase in complex2 with ram
|
|
|
|
| 9 | c3h96B_
|
|
 |
21.7 |
14 |
PDB header:flavoprotein
Chain: B: PDB Molecule:f420-h2 dependent reductase a;
PDBTitle: msmeg_3358 f420 reductase
|
|
|
|
| 10 | c5c6hN_
|
|
 |
20.3 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: N: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 11 | c5c6hT_
|
|
 |
20.3 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: T: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 12 | c5c6hH_
|
|
 |
20.3 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: H: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 13 | c5c6hD_
|
|
 |
20.3 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: D: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 14 | c5c6hP_
|
|
 |
20.3 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: P: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 15 | c5c6hB_
|
|
 |
19.9 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: B: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 16 | c5c6hX_
|
|
 |
19.6 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: X: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 17 | c3r5zB_
|
|
 |
18.8 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of a deazaflavin-dependent reductase from nocardia2 farcinica, with co-factor f420
|
|
|
|
| 18 | c5c6hR_
|
|
 |
18.1 |
42 |
PDB header:apoptosis/apoptosis regulator
Chain: R: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 19 | c5c6hL_
|
|
 |
16.5 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: L: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 20 | c5c6hF_
|
|
 |
15.8 |
47 |
PDB header:apoptosis/apoptosis regulator
Chain: F: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
|
| 21 | d1k99a_ |
|
not modelled |
13.4 |
7 |
Fold:HMG-box
Superfamily:HMG-box
Family:HMG-box |
|
|
| 22 | c5c6hV_ |
|
not modelled |
13.1 |
40 |
PDB header:apoptosis/apoptosis regulator
Chain: V: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
| 23 | c5c6hJ_ |
|
not modelled |
10.3 |
42 |
PDB header:apoptosis/apoptosis regulator
Chain: J: PDB Molecule:mule bh3 peptide from e3 ubiquitin-protein ligase huwe1;
PDBTitle: mcl-1 complexed with mule
|
|
|
| 24 | c1p58F_ |
|
not modelled |
9.7 |
23 |
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
|
|
|
| 25 | c6dftE_ |
|
not modelled |
9.6 |
29 |
PDB header:transferase
Chain: E: PDB Molecule:deoxyhypusine synthase;
PDBTitle: trypanosoma brucei deoxyhypusine synthase
|
|
|
| 26 | c5ht7A_ |
|
not modelled |
9.5 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a transition-metal-ion-binding betagamma-2 crystallin from methanosaeta thermophila
|
|
|
| 27 | c1p58E_ |
|
not modelled |
9.1 |
19 |
PDB header:virus
Chain: E: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
|
|
|
| 28 | c4lgdH_ |
|
not modelled |
9.0 |
26 |
PDB header:signaling protein
Chain: H: PDB Molecule:ras association domain family member 5, rassf5;
PDBTitle: structural basis for autoactivation of human mst2 kinase and its2 regulation by rassf5
|
|
|
| 29 | c5o60g_ |
|
not modelled |
8.2 |
50 |
PDB header:ribosome
Chain: G: PDB Molecule:50s ribosomal protein l6;
PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
|
|
|
| 30 | c6em5v_ |
|
not modelled |
8.0 |
31 |
PDB header:ribosome
Chain: V: PDB Molecule:60s ribosomal protein l23-a;
PDBTitle: state d architectural model (nsa1-tap flag-ytm1) - visualizing the2 assembly pathway of nucleolar pre-60s ribosomes
|
|
|
| 31 | c6dftJ_ |
|
not modelled |
7.8 |
31 |
PDB header:transferase
Chain: J: PDB Molecule:deoxyhypusine synthase regulatory subunit;
PDBTitle: trypanosoma brucei deoxyhypusine synthase
|
|
|
| 32 | d2i5ha1 |
|
not modelled |
7.8 |
22 |
Fold:AF1531-like
Superfamily:AF1531-like
Family:AF1531-like |
|
|
| 33 | c2i5hA_ |
|
not modelled |
7.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein af1531;
PDBTitle: crystal structure of af1531 from archaeoglobus fulgidus,2 pfam duf655
|
|
|
| 34 | c1geaA_ |
|
not modelled |
7.6 |
36 |
PDB header:neuropeptide
Chain: A: PDB Molecule:pituitary adenylate cyclase activating
PDBTitle: receptor-bound conformation of pacap21
|
|
|
| 35 | c1ckxA_ |
|
not modelled |
7.6 |
36 |
PDB header:metal transport
Chain: A: PDB Molecule:cystic fibrosis transmembrane conductance
PDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
|
|
|
| 36 | c4p63A_ |
|
not modelled |
6.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:probable deoxyhypusine synthase;
PDBTitle: crystal structure of deoxyhypusine synthase from pyrococcus horikoshii
|
|
|
| 37 | d2e74g1 |
|
not modelled |
6.6 |
29 |
Fold:Single transmembrane helix
Superfamily:PetG subunit of the cytochrome b6f complex
Family:PetG subunit of the cytochrome b6f complex |
|
|
| 38 | c1vf5G_ |
|
not modelled |
6.4 |
24 |
PDB header:photosynthesis
Chain: G: PDB Molecule:protein pet g;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
|
|
|
| 39 | d1vf5g_ |
|
not modelled |
6.4 |
24 |
Fold:Single transmembrane helix
Superfamily:PetG subunit of the cytochrome b6f complex
Family:PetG subunit of the cytochrome b6f complex |
|
|
| 40 | d1e7la1 |
|
not modelled |
6.3 |
38 |
Fold:LEM/SAP HeH motif
Superfamily:Recombination endonuclease VII, C-terminal and dimerization domains
Family:Recombination endonuclease VII, C-terminal and dimerization domains |
|
|
| 41 | c4bqqB_ |
|
not modelled |
6.3 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:integrase;
PDBTitle: protein crystal structure of the n-terminal and recombinase domains of2 the streptomyces temperate phage serine recombinase, fc31 integrase.
|
|
|
| 42 | c2mc0A_ |
|
not modelled |
6.2 |
23 |
PDB header:transcription activator/antibiotic
Chain: A: PDB Molecule:hth-type transcriptional activator tipa;
PDBTitle: structural basis of a thiopeptide antibiotic multidrug resistance2 system from streptomyces lividans:nosiheptide in complex with tipas
|
|
|
| 43 | c4hxhC_ |
|
not modelled |
5.8 |
22 |
PDB header:rna/rna binding protein/hydrolase
Chain: C: PDB Molecule:histone rna hairpin-binding protein;
PDBTitle: structure of mrna stem-loop, human stem-loop binding protein and2 3'hexo ternary complex
|
|
|
| 44 | d1dhsa_ |
|
not modelled |
5.8 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Deoxyhypusine synthase, DHS |
|
|
| 45 | c2yqrA_ |
|
not modelled |
5.6 |
20 |
PDB header:rna binding protein
Chain: A: PDB Molecule:kiaa0907 protein;
PDBTitle: solution structure of the kh domain in kiaa0907 protein
|
|
|
| 46 | c3j2pD_ |
|
not modelled |
5.5 |
21 |
PDB header:viral protein
Chain: D: PDB Molecule:small envelope protein m;
PDBTitle: cryoem structure of dengue virus envelope protein heterotetramer
|
|
|
| 47 | d2oy9a1 |
|
not modelled |
5.5 |
11 |
Fold:BH2638-like
Superfamily:BH2638-like
Family:BH2638-like |
|
|
| 48 | c5n7kD_ |
|
not modelled |
5.4 |
32 |
PDB header:cell adhesion
Chain: D: PDB Molecule:marvel domain-containing protein 2;
PDBTitle: crystal structure of the coiled-coil domain of human tricellulin
|
|
|
| 49 | c2na6C_ |
|
not modelled |
5.4 |
56 |
PDB header:apoptosis
Chain: C: PDB Molecule:tumor necrosis factor receptor superfamily member 6;
PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
|
|
|
| 50 | c2na6A_ |
|
not modelled |
5.4 |
56 |
PDB header:apoptosis
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily member 6;
PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
|
|
|
| 51 | d1hhsa_ |
|
not modelled |
5.4 |
44 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:dsRNA phage RNA-dependent RNA-polymerase |
|
|
| 52 | c2na6B_ |
|
not modelled |
5.4 |
56 |
PDB header:apoptosis
Chain: B: PDB Molecule:tumor necrosis factor receptor superfamily member 6;
PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
|
|
|