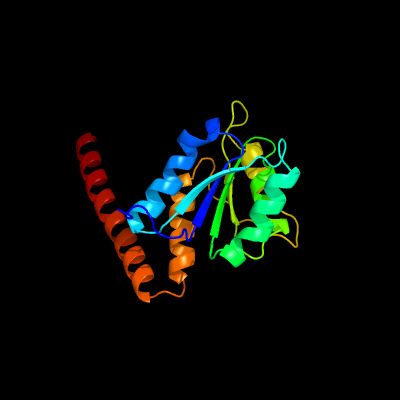
| 1 | c3lp6D_
|
|
 |
100.0 |
99 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
|
|
|
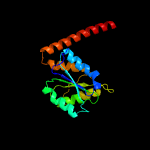
| 2 | d1o4va_
|
|
 |
100.0 |
58 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
|
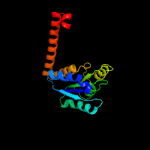
| 3 | c3trhI_
|
|
 |
100.0 |
41 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
|
|
|
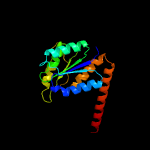
| 4 | c3orsD_
|
|
 |
100.0 |
57 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
|
|
|
| 5 | c4grdA_
|
|
 |
100.0 |
57 |
PDB header:lyase,isomerase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from burkholderia cenocepacia j2315
|
|
|
|
| 6 | c2fw9A_
|
|
 |
100.0 |
57 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
|
|
|
| 7 | d1u11a_
|
|
 |
100.0 |
58 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
|
| 8 | d1qcza_
|
|
 |
100.0 |
49 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
|
| 9 | c2ywxA_
|
|
 |
100.0 |
44 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
|
|
|
|
| 10 | c2h31A_
|
|
 |
100.0 |
25 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
|
|
|
| 11 | c3rggD_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
|
|
|
| 12 | c4b4kK_
|
|
 |
100.0 |
61 |
PDB header:isomerase
Chain: K: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of bacillus anthracis pure
|
|
|
|
| 13 | c6o55B_
|
|
 |
100.0 |
54 |
PDB header:isomerase
Chain: B: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 (pure) from legionella pneumophila
|
|
|
|
| 14 | d1xmpa_
|
|
 |
100.0 |
61 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
|
| 15 | c4ja0A_
|
|
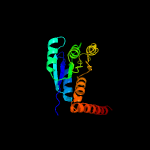 |
100.0 |
26 |
PDB header:protein binding
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase;
PDBTitle: crystal structure of the invertebrate bi-functional purine2 biosynthesis enzyme paics at 2.8 a resolution
|
|
|
|
| 16 | c5zxlD_
|
|
 |
98.1 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glycerol dehydrogenase;
PDBTitle: structure of glda from e.coli
|
|
|
|
| 17 | d1jq5a_
|
|
 |
98.0 |
19 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
|
| 18 | c4mcaB_
|
|
 |
97.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase from serratia to 1.9a
|
|
|
|
| 19 | c5tprB_
|
|
 |
97.8 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: desmethyl-4-deoxygadusol synthase from anabaena variabilis (ava_3858)2 with nad+ and zn2+ bound
|
|
|
|
| 20 | c6csjD_
|
|
 |
97.7 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:glycerol dehydrogenase;
PDBTitle: structure of a bacillus coagulans polyol dehydrogenase double mutant2 with an acquired d-lactate dehydrogenase activity
|
|
|
|
| 21 | c4p53A_ |
|
not modelled |
97.6 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:cyclase;
PDBTitle: vala (2-epi-5-epi-valiolone synthase) from streptomyces hygroscopicus2 subsp. jinggangensis 5008 with nad+ and zn2+ bound
|
|
|
| 22 | c3qbeA_ |
|
not modelled |
97.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of the 3-dehydroquinate synthase (arob) from2 mycobacterium tuberculosis
|
|
|
| 23 | c1ta9A_ |
|
not modelled |
97.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
|
|
|
| 24 | c3ce9A_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase (np_348253.1) from2 clostridium acetobutylicum at 2.37 a resolution
|
|
|
| 25 | c3okfA_ |
|
not modelled |
97.3 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
|
|
|
| 26 | d1o2da_ |
|
not modelled |
97.3 |
20 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 27 | c5eksB_ |
|
not modelled |
97.3 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: structure of 3-dehydroquinate synthase from acinetobacter baumannii in2 complex with nad
|
|
|
| 28 | d1vlja_ |
|
not modelled |
97.2 |
19 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 29 | c3ox4D_ |
|
not modelled |
97.2 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 2;
PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
|
|
|
| 30 | c3bfjK_ |
|
not modelled |
97.2 |
14 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:1,3-propanediol oxidoreductase;
PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
|
|
|
| 31 | c3zokB_ |
|
not modelled |
97.1 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: structure of 3-dehydroquinate synthase from actinidia chinensis in2 complex with nad
|
|
|
| 32 | c6c76A_ |
|
not modelled |
97.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: structure of iron containing alcohol dehydrogenase from thermococcus2 thioreducens in an orthorhombic crystal form
|
|
|
| 33 | c3jzdA_ |
|
not modelled |
96.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-containing alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehedrogenase (yp_298327.1) from2 ralstonia eutropha jmp134 at 2.10 a resolution
|
|
|
| 34 | d1rrma_ |
|
not modelled |
96.9 |
14 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 35 | c3zdrA_ |
|
not modelled |
96.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase domain of the bifunctional
PDBTitle: structure of the alcohol dehydrogenase (adh) domain of a2 bifunctional adhe dehydrogenase from geobacillus3 thermoglucosidasius ncimb 11955
|
|
|
| 36 | c3uhjE_ |
|
not modelled |
96.9 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:probable glycerol dehydrogenase;
PDBTitle: crystal structure of a probable glycerol dehydrogenase from2 sinorhizobium meliloti 1021
|
|
|
| 37 | c6c5cA_ |
|
not modelled |
96.9 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of aro1p from candida albicans sc5314 in complex2 with nadh
|
|
|
| 38 | c5yvmA_ |
|
not modelled |
96.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the archaeal halo-thermophilic red sea brine pool2 alcohol dehydrogenase adh/d1 bound to nzq
|
|
|
| 39 | c3iv7B_ |
|
not modelled |
96.7 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase iv;
PDBTitle: crystal structure of iron-containing alcohol dehydrogenase2 (np_602249.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.07 a resolution
|
|
|
| 40 | c3hl0B_ |
|
not modelled |
96.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:maleylacetate reductase;
PDBTitle: crystal structure of maleylacetate reductase from agrobacterium2 tumefaciens
|
|
|
| 41 | d1sg6a_ |
|
not modelled |
96.6 |
19 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Dehydroquinate synthase, DHQS |
|
|
| 42 | c4fr2A_ |
|
not modelled |
96.4 |
16 |
PDB header:oxidoreductase, metal binding protein
Chain: A: PDB Molecule:1,3-propanediol dehydrogenase;
PDBTitle: alcohol dehydrogenase from oenococcus oeni
|
|
|
| 43 | c5hvnA_ |
|
not modelled |
96.4 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 3.0 angstrom crystal structure of 3-dehydroquinate synthase (arob)2 from francisella tularensis in complex with nad.
|
|
|
| 44 | c2gruB_ |
|
not modelled |
96.4 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:2-deoxy-scyllo-inosose synthase;
PDBTitle: crystal structure of 2-deoxy-scyllo-inosose synthase2 complexed with carbaglucose-6-phosphate, nad+ and co2+
|
|
|
| 45 | c3clhA_ |
|
not modelled |
96.0 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of 3-dehydroquinate synthase (dhqs)from2 helicobacter pylori
|
|
|
| 46 | d1oj7a_ |
|
not modelled |
95.9 |
18 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 47 | c2q5cA_ |
|
not modelled |
95.6 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
|
|
| 48 | d2pjua1 |
|
not modelled |
95.5 |
17 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
|
|
| 49 | d1kq3a_ |
|
not modelled |
95.4 |
16 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 50 | c2bonB_ |
|
not modelled |
95.3 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:lipid kinase;
PDBTitle: structure of an escherichia coli lipid kinase (yegs)
|
|
|
| 51 | d2bona1 |
|
not modelled |
95.1 |
20 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 52 | c3rf7A_ |
|
not modelled |
95.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-containing alcohol dehydrogenase;
PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
|
|
|
| 53 | d1ujna_ |
|
not modelled |
94.6 |
21 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Dehydroquinate synthase, DHQS |
|
|
| 54 | c6jkpD_ |
|
not modelled |
94.2 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:methanol dehydrogenase;
PDBTitle: crystal structure of sulfoacetaldehyde reductase from bifidobacterium2 kashiwanohense in complex with nad+
|
|
|
| 55 | c4kqcA_ |
|
not modelled |
93.9 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: abc transporter, laci family transcriptional regulator from2 brachyspira murdochii
|
|
|
| 56 | d2jgra1 |
|
not modelled |
93.7 |
20 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 57 | d2p1ra1 |
|
not modelled |
93.7 |
20 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 58 | c4zhtB_ |
|
not modelled |
93.6 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:bifunctional udp-n-acetylglucosamine 2-epimerase/n-
PDBTitle: crystal structure of udp-glcnac 2-epimerase
|
|
|
| 59 | c2qq1A_ |
|
not modelled |
92.8 |
18 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdenum cofactor biosynthesis mog;
PDBTitle: crystal structure of molybdenum cofactor biosynthesis2 (aq_061) other form from aquifex aeolicus vf5
|
|
|
| 60 | d1guda_ |
|
not modelled |
92.8 |
14 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 61 | c4pe6B_ |
|
not modelled |
92.7 |
12 |
PDB header:solute-binding protein
Chain: B: PDB Molecule:putative abc transporter;
PDBTitle: crystal structure of abc transporter solute binding protein from2 thermobispora bispora dsm 43833
|
|
|
| 62 | c1xahA_ |
|
not modelled |
92.7 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: crystal structure of staphlyococcus aureus 3-dehydroquinate2 synthase (dhqs) in complex with zn2+ and nad+
|
|
|
| 63 | c5fb3C_ |
|
not modelled |
92.7 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerol-1-phosphate dehydrogenase [nad(p)+];
PDBTitle: structure of glycerophosphate dehydrogenase in complex with nadph
|
|
|
| 64 | d2qv7a1 |
|
not modelled |
92.5 |
20 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 65 | c3rotA_ |
|
not modelled |
92.0 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:abc sugar transporter, periplasmic sugar binding protein;
PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
|
|
|
| 66 | c3s99A_ |
|
not modelled |
91.6 |
12 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:basic membrane lipoprotein;
PDBTitle: crystal structure of a basic membrane lipoprotein from brucella2 melitensis, iodide soak
|
|
|
| 67 | c2qv7A_ |
|
not modelled |
91.2 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase dgkb;
PDBTitle: crystal structure of diacylglycerol kinase dgkb in complex with adp2 and mg
|
|
|
| 68 | c4irxA_ |
|
not modelled |
90.8 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:sugar abc transporter, periplasmic sugar-binding protein;
PDBTitle: crystal structure of caulobacter myo-inositol binding protein bound to2 myo-inositol
|
|
|
| 69 | c5z9aB_ |
|
not modelled |
90.8 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:chorismate synthase;
PDBTitle: crystal structure of chorismate synthase from pseudomonas aeruginosa
|
|
|
| 70 | c4z0nA_ |
|
not modelled |
90.8 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of a periplasmic solute binding protein (ipr025997)2 from streptobacillus moniliformis dsm-12112 (smon_0317, target efi-3 511281) with bound d-galactose
|
|
|
| 71 | c4werA_ |
|
not modelled |
90.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase catalytic domain protein;
PDBTitle: crystal structure of diacylglycerol kinase catalytic domain protein2 from enterococcus faecalis v583
|
|
|
| 72 | c5ulbA_ |
|
not modelled |
90.6 |
13 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative sugar abc transporter;
PDBTitle: crystal structure of sugar abc transporter from yersinia2 enterocolitica subsp. enterocolitica 8081
|
|
|
| 73 | c2pjuD_ |
|
not modelled |
90.5 |
17 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon regulatory protein2 prpr
|
|
|
| 74 | d1um0a_ |
|
not modelled |
90.2 |
23 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
|
|
| 75 | c3lftA_ |
|
not modelled |
90.0 |
16 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the abc domain in complex with l-trp from2 streptococcus pneumonia to 1.35a
|
|
|
| 76 | c5hsgA_ |
|
not modelled |
89.7 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter, nucleotide binding/atpase
PDBTitle: crystal structure of an abc transporter solute binding protein from2 klebsiella pneumoniae (kpn_01730, target efi-511059), apo open3 structure
|
|
|
| 77 | c2fn9A_ |
|
not modelled |
89.4 |
14 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:ribose abc transporter, periplasmic ribose-binding protein;
PDBTitle: thermotoga maritima ribose binding protein unliganded form
|
|
|
| 78 | c4rk5A_ |
|
not modelled |
89.4 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcriptional regulator from2 lactobacillus casei, target efi-512911, with bound sucrose
|
|
|
| 79 | c3u7rB_ |
|
not modelled |
89.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-dependent fmn reductase;
PDBTitle: ferb - flavoenzyme nad(p)h:(acceptor) oxidoreductase (ferb) from2 paracoccus denitrificans
|
|
|
| 80 | c5dkvD_ |
|
not modelled |
88.6 |
15 |
PDB header:sugar binding protein
Chain: D: PDB Molecule:abc transporter substrate binding protein (ribose);
PDBTitle: crystal structure of an abc transporter solute binding protein from2 agrobacterium vitis(avis_5339, target efi-511225) bound with alpha-d-3 tagatopyranose
|
|
|
| 81 | c4yleA_ |
|
not modelled |
88.5 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of an abc transpoter solute binding protein2 (ipr025997) from burkholderia multivorans (bmul_1631, target efi-3 511115) with an unknown ligand modelled as alpha-d-erythrofuranose
|
|
|
| 82 | c4ycsC_ |
|
not modelled |
88.5 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of putative lipoprotein from peptoclostridium2 difficile 630 (fragment)
|
|
|
| 83 | c4kvfA_ |
|
not modelled |
88.5 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:rhamnose abc transporter, periplasmic rhamnose-binding
PDBTitle: the crystal structure of a rhamnose abc transporter, periplasmic2 rhamnose-binding protein from kribbella flavida dsm 17836
|
|
|
| 84 | c5tt0A_ |
|
not modelled |
88.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: crystal structure of an oxidoreductase (short chain2 dehydrogenase/reductase family) from burkholderia thailandensis
|
|
|
| 85 | c4lj2A_ |
|
not modelled |
87.3 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:chorismate synthase;
PDBTitle: crystal structure of chorismate synthase from acinetobacter baumannii2 at 3.15a resolution
|
|
|
| 86 | c4rflB_ |
|
not modelled |
86.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-1-phosphate dehydrogenase [nad(p)+];
PDBTitle: crystal structure of g1pdh with nadph from methanocaldococcus2 jannaschii
|
|
|
| 87 | c5dteD_ |
|
not modelled |
86.5 |
19 |
PDB header:transport protein
Chain: D: PDB Molecule:monosaccharide-transporting atpase;
PDBTitle: crystal structure of an abc transporter periplasmic solute binding2 protein (ipr025997) from actinobacillus succinogenes 130z(asuc_0081,3 target efi-511065) with bound d-allose
|
|
|
| 88 | c3hcwB_ |
|
not modelled |
86.2 |
8 |
PDB header:rna binding protein
Chain: B: PDB Molecule:maltose operon transcriptional repressor;
PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
|
|
|
| 89 | d1q1la_ |
|
not modelled |
85.8 |
24 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
|
|
| 90 | c1zuwA_ |
|
not modelled |
85.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
|
|
| 91 | d1sq1a_ |
|
not modelled |
85.5 |
31 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
|
|
| 92 | c1ztbA_ |
|
not modelled |
85.3 |
26 |
PDB header:ligase
Chain: A: PDB Molecule:chorismate synthase;
PDBTitle: crystal structure of chorismate synthase from mycobacterium2 tuberculosis
|
|
|
| 93 | d1qxoa_ |
|
not modelled |
84.7 |
30 |
Fold:Chorismate synthase, AroC
Superfamily:Chorismate synthase, AroC
Family:Chorismate synthase, AroC |
|
|
| 94 | c2fqxA_ |
|
not modelled |
84.5 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:membrane lipoprotein tmpc;
PDBTitle: pnra from treponema pallidum complexed with guanosine
|
|
|
| 95 | c4rxuA_ |
|
not modelled |
84.5 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic sugar-binding protein;
PDBTitle: crystal structure of carbohydrate transporter solute binding protein2 caur_1924 from chloroflexus aurantiacus, target efi-511158, in3 complex with d-glucose
|
|
|
| 96 | c3h75A_ |
|
not modelled |
84.2 |
18 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic sugar-binding domain protein;
PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
|
|
|
| 97 | c3h8qB_ |
|
not modelled |
84.0 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 3;
PDBTitle: crystal structure of glutaredoxin domain of human thioredoxin2 reductase 3
|
|
|
| 98 | d1xi8a3 |
|
not modelled |
83.9 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 99 | c4trrH_ |
|
not modelled |
83.6 |
20 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:putative d-beta-hydroxybutyrate dehydrogenase;
PDBTitle: crystal structure of a putative putative d-beta-hydroxybutyrate2 dehydrogenase from burkholderia cenocepacia j2315
|
|
|
| 100 | d2gm3a1 |
|
not modelled |
83.6 |
10 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
|
|
| 101 | c2jfoB_ |
|
not modelled |
83.4 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
|
|
| 102 | c4y9tA_ |
|
not modelled |
83.1 |
18 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:abc transporter, solute binding protein;
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from agrobacterium vitis s4 (avi_5305, target efi-511224)3 with bound alpha-d-glucosamine
|
|
|
| 103 | c5w16D_ |
|
not modelled |
82.9 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from thermus thermophilus in2 complex with d-glutamate
|
|
|
| 104 | c3opyH_ |
|
not modelled |
82.8 |
18 |
PDB header:transferase
Chain: H: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 105 | c3opyB_ |
|
not modelled |
82.8 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
|
|
| 106 | c2iy3A_ |
|
not modelled |
82.8 |
20 |
PDB header:rna-binding
Chain: A: PDB Molecule:signal recognition particle protein,signal recognition
PDBTitle: structure of the e. coli signal regognition particle
|
|
|
| 107 | c4rxmA_ |
|
not modelled |
82.5 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:possible sugar abc superfamily atp binding cassette
PDBTitle: crystal structure of periplasmic abc transporter solute binding2 protein a7jw62 from mannheimia haemolytica phl213, target efi-511105,3 in complex with myo-inositol
|
|
|
| 108 | c3ksmA_ |
|
not modelled |
82.4 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
|
|
| 109 | c2f59B_ |
|
not modelled |
82.2 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:6,7-dimethyl-8-ribityllumazine synthase 1;
PDBTitle: lumazine synthase ribh1 from brucella abortus (gene bruab1_0785,2 swiss-prot entry q57dy1) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
|
|
|
| 110 | c4pevB_ |
|
not modelled |
81.3 |
14 |
PDB header:solute-binding protein
Chain: B: PDB Molecule:membrane lipoprotein family protein;
PDBTitle: crystal structure of abc transporter system solute-binding proteins2 from aeropyrum pernix k1
|
|
|
| 111 | c5zvlB_ |
|
not modelled |
81.2 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutaredoxin;
PDBTitle: crystal structure of wheat glutarredoxin
|
|
|
| 112 | c3ak4C_ |
|
not modelled |
81.1 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-dependent quinuclidinone reductase;
PDBTitle: crystal structure of nadh-dependent quinuclidinone reductase from2 agrobacterium tumefaciens
|
|
|
| 113 | c1z0zC_ |
|
not modelled |
81.1 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:probable inorganic polyphosphate/atp-nad kinase;
PDBTitle: crystal structure of a nad kinase from archaeoglobus2 fulgidus in complex with nad
|
|
|
| 114 | c4o5aA_ |
|
not modelled |
80.7 |
16 |
PDB header:transcription regulator
Chain: A: PDB Molecule:laci family transcription regulator;
PDBTitle: the crystal structure of a laci family transcriptional regulator from2 bifidobacterium animalis subsp. lactis dsm 10140
|
|
|
| 115 | c3brsA_ |
|
not modelled |
80.7 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of sugar transporter from clostridium2 phytofermentans
|
|
|
| 116 | c3s40C_ |
|
not modelled |
80.6 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
|
|
| 117 | c3h7aC_ |
|
not modelled |
79.9 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of short-chain dehydrogenase from rhodopseudomonas2 palustris
|
|
|
| 118 | d2f7wa1 |
|
not modelled |
79.7 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 119 | d1z0sa1 |
|
not modelled |
79.5 |
16 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:NAD kinase-like |
|
|
| 120 | c3l49D_ |
|
not modelled |
78.7 |
14 |
PDB header:transport protein
Chain: D: PDB Molecule:abc sugar (ribose) transporter, periplasmic substrate-
PDBTitle: crystal structure of abc sugar transporter subunit from rhodobacter2 sphaeroides 2.4.1
|
|
|