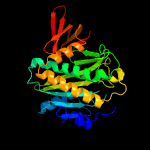
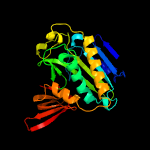
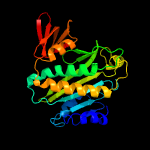
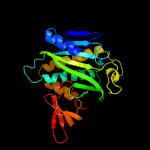
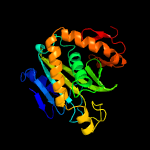
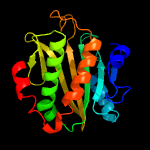
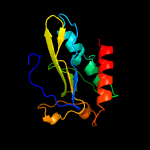
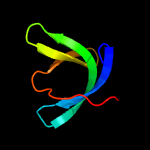
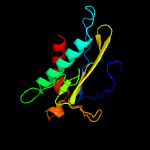
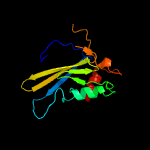
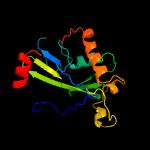
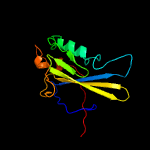
1 c2ewnA_
100.0
33
PDB header: ligase, transcriptionChain: A: PDB Molecule: bira bifunctional protein;PDBTitle: ecoli biotin repressor with co-repressor analog
2 c2cghB_
100.0
100
PDB header: ligaseChain: B: PDB Molecule: biotin ligase;PDBTitle: crystal structure of biotin ligase from mycobacterium tuberculosis
3 c2ej9A_
100.0
25
PDB header: ligaseChain: A: PDB Molecule: putative biotin ligase;PDBTitle: crystal structure of biotin protein ligase from2 methanococcus jannaschii
4 c3rkxA_
100.0
23
PDB header: ligaseChain: A: PDB Molecule: biotin-[acetyl-coa-carboxylase] ligase;PDBTitle: structural characterisation of staphylococcus aureus biotin protein2 ligase
5 c2eayB_
100.0
24
PDB header: ligaseChain: B: PDB Molecule: biotin [acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
6 c3bfmA_
100.0
15
PDB header: unknown functionChain: A: PDB Molecule: biotin protein ligase-like protein of unknown function;PDBTitle: crystal structure of a biotin protein ligase-like protein of unknown2 function (tm1040_0394) from silicibacter sp. tm1040 at 1.70 a3 resolution
7 c2dzcA_
100.0
28
PDB header: ligaseChain: A: PDB Molecule: biotin--[acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from pyrococcus2 horikoshii, mutation r48a
8 d2zgwa2
100.0
23
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Biotin holoenzyme synthetase
9 c6ck0A_
100.0
23
PDB header: ligaseChain: A: PDB Molecule: biotin acetyl coenzyme a carboxylase synthetase;PDBTitle: crystal structure of biotin acetyl coenzyme a carboxylase synthetase2 from helicobacter pylori with bound biotinylated atp
10 d1biaa3
100.0
32
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Biotin holoenzyme synthetase
11 c2qhvA_
98.8
16
PDB header: transferaseChain: A: PDB Molecule: lipoyltransferase;PDBTitle: structural basis of octanoic acid recognition by lipoate-protein2 ligase b
12 c2qhsA_
98.8
16
PDB header: transferaseChain: A: PDB Molecule: lipoyltransferase;PDBTitle: structural basis of octanoic acid recognition by lipoate-protein2 ligase b
13 d1biaa2
98.7
34
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)
14 d1w66a1
98.6
16
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
15 c1vqzA_
98.1
12
PDB header: ligaseChain: A: PDB Molecule: lipoate-protein ligase, putative;PDBTitle: crystal structure of a putative lipoate-protein ligase a (sp_1160)2 from streptococcus pneumoniae tigr4 at 1.99 a resolution
16 d2p0la1
98.0
19
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
17 d1vqza2
98.0
14
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
18 c3r07A_
97.9
16
PDB header: transferaseChain: A: PDB Molecule: lipoate-protein ligase a subunit 1;PDBTitle: structural analysis of an archaeal lipoylation system. a bi-partite2 lipoate protein ligase and its e2 lipoyl domain from thermoplasma3 acidophilum
19 d2c8ma1
97.8
16
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
20 d1x2ga2
97.6
13
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
21 d2zgwa1
not modelled
97.4
50
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)
22 c2e5aA_
not modelled
97.3
12
PDB header: ligaseChain: A: PDB Molecule: lipoyltransferase 1;PDBTitle: crystal structure of bovine lipoyltransferase in complex2 with lipoyl-amp
23 c5ij6A_
not modelled
97.1
18
PDB header: ligase,transferaseChain: A: PDB Molecule: lipoate--protein ligase;PDBTitle: crystal structure of enterococcus faecalis lipoate-protein ligase a2 (lpla-1) in complex with lipoic acid
24 c5ibyA_
not modelled
97.0
13
PDB header: ligase,transferaseChain: A: PDB Molecule: lipoate--protein ligase;PDBTitle: crystal structure of enterococcus faecalis lipoate-protein ligase a2 (lpla-2) in complex with lipoic acid
25 d2p5ia1
not modelled
97.0
18
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: LplA-like
26 c1x2gB_
not modelled
96.6
14
PDB header: ligaseChain: B: PDB Molecule: lipoate-protein ligase a;PDBTitle: crystal structure of lipate-protein ligase a from2 escherichia coli
27 c5t8uA_
not modelled
94.4
13
PDB header: ligaseChain: A: PDB Molecule: lipoate-protein ligase 1;PDBTitle: crystal structure of p. falciparum lipl1 in complex lipoate
28 d1d3bb_
not modelled
92.9
20
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
29 d1i8fa_
not modelled
92.8
31
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
30 d1d3bl_
not modelled
92.6
20
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
31 c4c92G_
not modelled
92.4
36
PDB header: transcriptionChain: G: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: crystal structure of the yeast lsm1-7 complex
32 c3swnC_
not modelled
92.4
27
PDB header: rna binding proteinChain: C: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
33 c4m77H_
not modelled
91.5
21
PDB header: structural proteinChain: H: PDB Molecule: u6 snrna-associated sm-like protein lsm8;PDBTitle: crystal structure of lsm2-8 complex, space group i212121
34 c3jb9b_
not modelled
91.4
18
PDB header: rna binding protein/rnaChain: B: PDB Molecule: pre-mrna-splicing factor cwf10;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
35 c3jcr7_
not modelled
91.2
32
PDB header: splicingChain: 7: PDB Molecule: lsm7;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
36 c3jb9E_
not modelled
91.2
19
PDB header: rna binding protein/rnaChain: E: PDB Molecule: small nuclear ribonucleoprotein-associated protein b;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
37 c4m78H_
not modelled
91.2
21
PDB header: rna binding proteinChain: H: PDB Molecule: u6 snrna-associated sm-like protein lsm8;PDBTitle: crystal structure of lsm2-8 complex, space group p21
38 d2ddza1
not modelled
91.1
18
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: PH0223-like
39 c3jcr5_
not modelled
90.8
27
PDB header: splicingChain: 5: PDB Molecule: lsm5;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
40 c2fwkB_
not modelled
90.7
30
PDB header: dna binding proteinChain: B: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: crystal structure of cryptosporidium parvum u6 snrna-associated sm-2 like protein lsm5
41 c3jb9J_
not modelled
90.7
27
PDB header: rna binding protein/rnaChain: J: PDB Molecule: small nuclear ribonucleoprotein g;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
42 c4c8qE_
not modelled
90.6
24
PDB header: transcriptionChain: E: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: crystal structure of the yeast lsm1-7-pat1 complex
43 d1h641_
not modelled
90.6
21
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
44 c5mknK_
not modelled
90.5
27
PDB header: rna binding proteinChain: K: PDB Molecule: like-sm ribonucleoprotein core;PDBTitle: crystal structure of smap (lsm) protein from methanococcus vannielii
45 c3cw15_
not modelled
90.5
27
PDB header: splicingChain: 5: PDB Molecule: small nuclear ribonucleoprotein g;PDB Fragment: residues 1-215;
PDBTitle: crystal structure of human spliceosomal u1 snrnp
46 d1th7a1
not modelled
90.4
30
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
47 c3jcr8_
not modelled
90.1
30
PDB header: splicingChain: 8: PDB Molecule: lsm8;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
48 c3swnA_
not modelled
90.1
24
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
49 c3swnT_
not modelled
90.0
29
PDB header: rna binding proteinChain: T: PDB Molecule: u6 snrna-associated sm-like protein lsm6;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
50 d1m5q1_
not modelled
89.9
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
51 c3qiiA_
not modelled
89.8
13
PDB header: transcription regulatorChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 2 of human phd finger protein 20
52 c3bw1A_
not modelled
89.8
24
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm3;PDBTitle: crystal structure of homomeric yeast lsm3 exhibiting novel octameric2 ring organisation
53 d1mgqa_
not modelled
89.8
24
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
54 c3cw1Z_
not modelled
89.8
23
PDB header: splicingChain: Z: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of human spliceosomal u1 snrnp
55 c4emgM_
not modelled
89.6
21
PDB header: rna binding proteinChain: M: PDB Molecule: probable u6 snrna-associated sm-like protein lsm3;PDBTitle: crystal structure of splsm3
56 d1i4k1_
not modelled
89.6
30
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
57 c4xq3F_
not modelled
89.4
27
PDB header: rna binding proteinChain: F: PDB Molecule: like-sm ribonucleoprotein core;PDBTitle: crystal structure of sso-smap2
58 c3pgwB_
not modelled
89.0
24
PDB header: splicing/dna/rnaChain: B: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
59 c4c8qF_
not modelled
88.8
30
PDB header: transcriptionChain: F: PDB Molecule: u6 snrna-associated sm-like protein lsm6;PDBTitle: crystal structure of the yeast lsm1-7-pat1 complex
60 c3jcr3_
not modelled
88.2
15
PDB header: splicingChain: 3: PDB Molecule: lsm3;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
61 c4c8qA_
not modelled
88.0
23
PDB header: transcriptionChain: A: PDB Molecule: sm-like protein lsm1;PDBTitle: crystal structure of the yeast lsm1-7-pat1 complex
62 d1d3ba_
not modelled
87.9
18
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
63 c3pgwQ_
not modelled
87.9
24
PDB header: splicing/dna/rnaChain: Q: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
64 c1b34A_
not modelled
87.7
23
PDB header: rna binding proteinChain: A: PDB Molecule: protein (small nuclear ribonucleoprotein sm d1);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
65 d1b34a_
not modelled
87.7
23
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
66 d1jbma_
not modelled
87.4
25
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
67 c5ganr_
not modelled
87.2
20
PDB header: transcriptionChain: R: PDB Molecule: PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
68 c1n9sH_
not modelled
87.0
24
PDB header: translationChain: H: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of yeast smf in spacegroup p43212
69 c4c92D_
not modelled
86.9
27
PDB header: transcriptionChain: D: PDB Molecule: u6 snrna-associated sm-like protein lsm4;PDBTitle: crystal structure of the yeast lsm1-7 complex
70 c3jb9D_
not modelled
86.6
24
PDB header: rna binding protein/rnaChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
71 c2ldmA_
not modelled
85.9
13
PDB header: transcription/protein bindingChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of human phf20 tudor2 domain bound to a p53 segment2 containing a dimethyllysine analog p53k370me2
72 c4c92A_
not modelled
85.7
23
PDB header: transcriptionChain: A: PDB Molecule: sm-like protein lsm1;PDBTitle: crystal structure of the yeast lsm1-7 complex
73 d2fwka1
not modelled
85.7
30
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
74 c5gaon_
not modelled
85.6
24
PDB header: transcriptionChain: N: PDB Molecule: PDBTitle: head region of the yeast spliceosomal u4/u6.u5 tri-snrnp
75 c3cw1A_
not modelled
85.4
27
PDB header: splicingChain: A: PDB Molecule: small nuclear ribonucleoprotein-associated proteins b andPDBTitle: crystal structure of human spliceosomal u1 snrnp
76 c5ganm_
not modelled
85.3
10
PDB header: transcriptionChain: M: PDB Molecule: PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
77 c3jcr4_
not modelled
84.5
21
PDB header: splicingChain: 4: PDB Molecule: lsm4;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
78 c4uc4A_
not modelled
84.4
17
PDB header: replicationChain: A: PDB Molecule: lysine-specific demethylase 4b;PDBTitle: crystal structure of hybrid tudor domain of human lysine demethylase2 kdm4b
79 c2qqsB_
not modelled
84.2
23
PDB header: oxidoreductaseChain: B: PDB Molecule: jmjc domain-containing histone demethylationPDBTitle: jmjd2a tandem tudor domains in complex with a trimethylated2 histone h4-k20 peptide
80 c3jcr2_
not modelled
82.8
24
PDB header: splicingChain: 2: PDB Molecule: lsm2;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
81 c4c92B_
not modelled
82.8
27
PDB header: transcriptionChain: B: PDB Molecule: u6 snrna-associated sm-like protein lsm2;PDBTitle: crystal structure of the yeast lsm1-7 complex
82 c3p8dB_
not modelled
82.8
13
PDB header: protein bindingChain: B: PDB Molecule: medulloblastoma antigen mu-mb-50.72;PDBTitle: crystal structure of the second tudor domain of human phf20 (homodimer2 form)
83 c5gank_
not modelled
82.2
24
PDB header: transcriptionChain: K: PDB Molecule: 13 kda ribonucleoprotein-associated protein;PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
84 c4emhY_
not modelled
82.2
29
PDB header: rna binding proteinChain: Y: PDB Molecule: PDBTitle: crystal structure of splsm4
85 c3cw1D_
not modelled
81.3
18
PDB header: splicingChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: crystal structure of human spliceosomal u1 snrnp
86 d1n9sc_
not modelled
81.3
24
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
87 c5uz5K_
not modelled
81.1
26
PDB header: nuclear protein/rnaChain: K: PDB Molecule: small nuclear ribonucleoprotein-associated protein b;PDBTitle: s. cerevisiae u1 snrnp
88 d1n9ra_
not modelled
80.2
21
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
89 d1ljoa_
not modelled
79.9
35
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
90 c1b34B_
not modelled
79.8
13
PDB header: rna binding proteinChain: B: PDB Molecule: protein (small nuclear ribonucleoprotein sm d2);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
91 d1b34b_
not modelled
79.8
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP
92 c2equA_
not modelled
76.1
11
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
93 c2e5pA_
not modelled
72.7
16
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 1 (phf1 protein)
94 c2xdpA_
not modelled
72.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 4c;PDBTitle: crystal structure of the tudor domain of human jmjd2c
95 c3jcr6_
not modelled
68.9
30
PDB header: splicingChain: 6: PDB Molecule: lsm6;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
96 c5gaol_
not modelled
64.1
24
PDB header: transcriptionChain: L: PDB Molecule: PDBTitle: head region of the yeast spliceosomal u4/u6.u5 tri-snrnp
97 d1sg5a1
not modelled
62.7
19
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: Rof-like
98 c3jb9m_
not modelled
62.5
11
PDB header: rna binding protein/rnaChain: M: PDB Molecule: pre-mrna-processing protein 45;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
99 c3cw1W_
not modelled
60.6
19
PDB header: splicingChain: W: PDB Molecule: small nuclear ribonucleoprotein e;PDBTitle: crystal structure of human spliceosomal u1 snrnp
100 c5ganp_
not modelled
55.5
9
PDB header: transcriptionChain: P: PDB Molecule: PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
101 c1xniI_
not modelled
55.4
22
PDB header: cell cycleChain: I: PDB Molecule: tumor suppressor p53-binding protein 1;PDBTitle: tandem tudor domain of 53bp1
102 c2e5qA_
not modelled
54.5
18
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 19;PDBTitle: solution structure of the tudor domain of phd finger2 protein 19, isoform b [homo sapiens]
103 c3pgwV_
not modelled
51.7
11
PDB header: splicing/dna/rnaChain: V: PDB Molecule: sm-d2;PDBTitle: crystal structure of human u1 snrnp
104 c5gl6A_
not modelled
49.7
7
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome maturation factor rimp;PDBTitle: msmeg rimp
105 d1ib8a1
not modelled
48.7
15
Fold: Sm-like foldSuperfamily: YhbC-like, C-terminal domainFamily: YhbC-like, C-terminal domain
106 c4qtkB_
not modelled
46.6
21
PDB header: transcription/dnaChain: B: PDB Molecule: white-opaque regulator 1;PDBTitle: complex of wopr domain of wor1 in candida albicans with the 17bp dsdna
107 c4a53A_
not modelled
45.2
25
PDB header: rna binding proteinChain: A: PDB Molecule: edc3;PDBTitle: structural basis of the dcp1:dcp2 mrna decapping complex activation2 by edc3 and scd6
108 c4m8bR_
not modelled
45.0
20
PDB header: transcription/dnaChain: R: PDB Molecule: yhr177w;PDBTitle: fungal protein
109 c5szeA_
not modelled
44.6
19
PDB header: rna-binding protein/rnaChain: A: PDB Molecule: rna-binding protein hfq;PDBTitle: crystal structure of aquifex aeolicus hfq-rna complex at 1.5a
110 c4tseA_
not modelled
43.6
21
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase mib1;PDBTitle: crystal structure of the mib repeat domain of mind bomb 1
111 d2qqra2
not modelled
42.1
23
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain
112 c1y96D_
not modelled
41.7
19
PDB header: rna binding proteinChain: D: PDB Molecule: gem-associated protein 7;PDBTitle: crystal structure of the gemin6/gemin7 heterodimer from the2 human smn complex
113 c2eqjA_
not modelled
40.7
15
PDB header: transcriptionChain: A: PDB Molecule: metal-response element-binding transcriptionPDBTitle: solution structure of the tudor domain of metal-response2 element-binding transcription factor 2
114 c2m0oA_
not modelled
38.3
17
PDB header: peptide binding proteinChain: A: PDB Molecule: phd finger protein 1;PDBTitle: the solution structure of human phf1 in complex with h3k36me3
115 c1ssfA_
not modelled
37.5
20
PDB header: cell cycleChain: A: PDB Molecule: transformation related protein 53 bindingPDBTitle: solution structure of the mouse 53bp1 fragment (residues2 1463-1617)
116 c4a54A_
not modelled
33.6
27
PDB header: rna binding protein/hydrolaseChain: A: PDB Molecule: edc3;PDBTitle: structural basis of the dcp1:dcp2 mrna decapping complex activation2 by edc3 and scd6
117 c2hqxB_
not modelled
26.9
10
PDB header: transcriptionChain: B: PDB Molecule: p100 co-activator tudor domain;PDBTitle: crystal structure of human p100 tudor domain conserved2 region
118 d2hqxa1
not modelled
26.9
10
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain
119 c3lv2A_
not modelled
26.6
20
PDB header: transferaseChain: A: PDB Molecule: adenosylmethionine-8-amino-7-oxononanoate aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis 7,8-diaminopelargonic2 acid synthase in complex with substrate analog sinefungin
120 c6ekvA_
not modelled
25.5
44
PDB header: lipid binding proteinChain: A: PDB Molecule: toxin complex component orf-x2;PDBTitle: structure of orfx2 from clostridium botulinum a2